+Search query
-Structure paper
| Title | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 83, Issue 17, Page 3171-33187.e7, Year 2023 |
| Publish date | Sep 7, 2023 |
 Authors Authors | Chang Zhao / Heli Wang / Ying Liu / Lin Cheng / Bo Wang / Xiaowen Tian / Hong Fu / Chao Wu / Ziyan Li / Chenglong Shen / Jingjing Yu / Shengyong Yang / Hongbo Hu / Ping Fu / Liang Ma / Chuanxin Wang / Wei Yan / Zhenhua Shao /  |
| PubMed Abstract | Hydroxycarboxylic acid receptor 2 (HCAR2), modulated by endogenous ketone body β-hydroxybutyrate and exogenous niacin, is a promising therapeutic target for inflammation-related diseases. HCAR2 ...Hydroxycarboxylic acid receptor 2 (HCAR2), modulated by endogenous ketone body β-hydroxybutyrate and exogenous niacin, is a promising therapeutic target for inflammation-related diseases. HCAR2 mediates distinct pathophysiological events by activating G protein or β-arrestin effectors. Here, we characterize compound 9n as a G-biased allosteric modulator (BAM) of HCAR2 and exhibit anti-inflammatory efficacy in RAW264.7 macrophages via a specific HCAR2-G pathway. Furthermore, four structures of HCAR2-G complex bound to orthosteric agonists (niacin or monomethyl fumarate), compound 9n, and niacin together with compound 9n simultaneously reveal a common orthosteric site and a unique allosteric site. Combined with functional studies, we decipher the action framework of biased allosteric modulation of compound 9n on the orthosteric site. Moreover, co-administration of compound 9n with orthosteric agonists could enhance anti-inflammatory effects in the mouse model of colitis. Together, our study provides insight to understand the molecular pharmacology of the BAM and facilitates exploring the therapeutic potential of the BAM with orthosteric drugs. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:37597514 PubMed:37597514 |
| Methods | EM (single particle) |
| Resolution | 2.78 - 3.5 Å |
| Structure data | EMDB-36300, PDB-8jhy: EMDB-36312, PDB-8jii: EMDB-36317, PDB-8jil: EMDB-36318, PDB-8jim:  EMDB-36490: Cryo-EM map of human receptor R2  EMDB-36491: Cryo-EM map of human Gi heterotrimer  EMDB-36492: Cryo-EM map of human receptor R2  EMDB-36493: Cryo-EM map of human Gi heterotrimer  EMDB-36494: Cryo-EM map of human receptor R2  EMDB-36495: Cryo-EM map of human Gi heterotrimer  EMDB-36496: Cryo-EM map of human receptor R2  EMDB-36497: Cryo-EM map of human Gi heterotrimer  EMDB-36505: Cryo-EM map of human receptor R2 in complex with Gi protein  EMDB-36506: Cryo-EM map of human receptor R2 in complex with Gi protein  EMDB-36507: Cryo-EM map of human receptor R2 in complex with Gi protein  EMDB-36508: Cryo-EM map of human receptor R2 in complex with Gi protein |
| Chemicals | 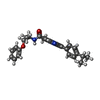 ChemComp-IX8:  ChemComp-CLR:  ChemComp-NIO:  ChemComp-HOH:  ChemComp-AW9: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  Complex / Complex /  Agonist Agonist |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













