+Search query
-Structure paper
| Title | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Year 2024 |
| Publish date | Feb 22, 2024 |
 Authors Authors | Kexin Hu / Qing Zhang / Yang Chen / Jintong Yang / Ying Xia / Bing Rao / Shaobai Li / Yafeng Shen / Mi Cao / Hongliang Lu / An Qin / Xian-Cheng Jiang / Deqiang Yao / Jie Zhao / Lu Zhou / Yu Cao /   |
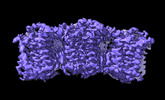
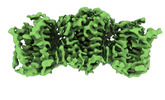
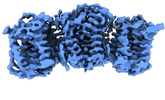
| PubMed Abstract | Sphingomyelin (SM) has key roles in modulating mammalian membrane properties and serves as an important pool for bioactive molecules. SM biosynthesis is mediated by the sphingomyelin synthase (SMS) ...Sphingomyelin (SM) has key roles in modulating mammalian membrane properties and serves as an important pool for bioactive molecules. SM biosynthesis is mediated by the sphingomyelin synthase (SMS) family, comprising SMS1, SMS2 and SMS-related (SMSr) members. Although SMS1 and SMS2 exhibit SMS activity, SMSr possesses ceramide phosphoethanolamine synthase activity. Here we determined the cryo-electron microscopic structures of human SMSr in complexes with ceramide, diacylglycerol/phosphoethanolamine and ceramide/phosphoethanolamine (CPE). The structures revealed a hexameric arrangement with a reaction chamber located between the transmembrane helices. Within this structure, a catalytic pentad E-H/D-H-D was identified, situated at the interface between the lipophilic and hydrophilic segments of the reaction chamber. Additionally, the study unveiled the two-step synthesis process catalyzed by SMSr, involving PE-PLC (phosphatidylethanolamine-phospholipase C) hydrolysis and the subsequent transfer of the phosphoethanolamine moiety to ceramide. This research provides insights into the catalytic mechanism of SMSr and expands our understanding of sphingolipid metabolism. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:38388831 PubMed:38388831 |
| Methods | EM (single particle) |
| Resolution | 3.29 - 3.74 Å |
| Structure data | EMDB-35492, PDB-8ijq: EMDB-35493, PDB-8ijr: EMDB-37383, PDB-8w9w: EMDB-37385, PDB-8w9y: |
| Chemicals |  ChemComp-16C: 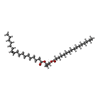 ChemComp-Z0P:  ChemComp-OPE: 
ChemComp-UJO: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  synthase / synthase /  sphingomyelin / CPE / sphingomyelin / CPE /  lipid metabolism lipid metabolism |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers