[English] 日本語
 Yorodumi
Yorodumi- EMDB-35492: The cryo-EM structure of human sphingomyelin synthase-related pro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 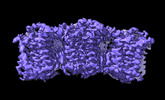 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  synthase / synthase /  sphingomyelin / CPE / sphingomyelin / CPE /  membrane protein / membrane protein /  lipid metabolism lipid metabolism | |||||||||
| Function / homology |  Function and homology information Function and homology informationceramide phosphoethanolamine biosynthetic process /  ceramide phosphoethanolamine synthase activity / ceramide phosphoethanolamine synthase activity /  sphingomyelin synthase activity / sphingomyelin synthase activity /  ceramide cholinephosphotransferase activity / regulation of ceramide biosynthetic process / ceramide cholinephosphotransferase activity / regulation of ceramide biosynthetic process /  Transferases; Transferring phosphorus-containing groups; Transferases for other substituted phosphate groups / sphingomyelin biosynthetic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / ceramide biosynthetic process ...ceramide phosphoethanolamine biosynthetic process / Transferases; Transferring phosphorus-containing groups; Transferases for other substituted phosphate groups / sphingomyelin biosynthetic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / ceramide biosynthetic process ...ceramide phosphoethanolamine biosynthetic process /  ceramide phosphoethanolamine synthase activity / ceramide phosphoethanolamine synthase activity /  sphingomyelin synthase activity / sphingomyelin synthase activity /  ceramide cholinephosphotransferase activity / regulation of ceramide biosynthetic process / ceramide cholinephosphotransferase activity / regulation of ceramide biosynthetic process /  Transferases; Transferring phosphorus-containing groups; Transferases for other substituted phosphate groups / sphingomyelin biosynthetic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / ceramide biosynthetic process / endoplasmic reticulum membrane / Transferases; Transferring phosphorus-containing groups; Transferases for other substituted phosphate groups / sphingomyelin biosynthetic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / ceramide biosynthetic process / endoplasmic reticulum membrane /  endoplasmic reticulum / endoplasmic reticulum /  membrane / membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.45 Å cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Hu K / Zhang Q / Chen Y / Yao D / Zhou L / Cao Y | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis. Authors: Kexin Hu / Qing Zhang / Yang Chen / Jintong Yang / Ying Xia / Bing Rao / Shaobai Li / Yafeng Shen / Mi Cao / Hongliang Lu / An Qin / Xian-Cheng Jiang / Deqiang Yao / Jie Zhao / Lu Zhou / Yu Cao /   Abstract: Sphingomyelin (SM) has key roles in modulating mammalian membrane properties and serves as an important pool for bioactive molecules. SM biosynthesis is mediated by the sphingomyelin synthase (SMS) ...Sphingomyelin (SM) has key roles in modulating mammalian membrane properties and serves as an important pool for bioactive molecules. SM biosynthesis is mediated by the sphingomyelin synthase (SMS) family, comprising SMS1, SMS2 and SMS-related (SMSr) members. Although SMS1 and SMS2 exhibit SMS activity, SMSr possesses ceramide phosphoethanolamine synthase activity. Here we determined the cryo-electron microscopic structures of human SMSr in complexes with ceramide, diacylglycerol/phosphoethanolamine and ceramide/phosphoethanolamine (CPE). The structures revealed a hexameric arrangement with a reaction chamber located between the transmembrane helices. Within this structure, a catalytic pentad E-H/D-H-D was identified, situated at the interface between the lipophilic and hydrophilic segments of the reaction chamber. Additionally, the study unveiled the two-step synthesis process catalyzed by SMSr, involving PE-PLC (phosphatidylethanolamine-phospholipase C) hydrolysis and the subsequent transfer of the phosphoethanolamine moiety to ceramide. This research provides insights into the catalytic mechanism of SMSr and expands our understanding of sphingolipid metabolism. | |||||||||
| History |
|
- Structure visualization
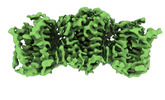
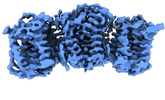
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35492.map.gz emd_35492.map.gz | 93.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35492-v30.xml emd-35492-v30.xml emd-35492.xml emd-35492.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35492_fsc.xml emd_35492_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_35492.png emd_35492.png | 96 KB | ||
| Filedesc metadata |  emd-35492.cif.gz emd-35492.cif.gz | 5.4 KB | ||
| Others |  emd_35492_half_map_1.map.gz emd_35492_half_map_1.map.gz emd_35492_half_map_2.map.gz emd_35492_half_map_2.map.gz | 91.6 MB 91.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35492 http://ftp.pdbj.org/pub/emdb/structures/EMD-35492 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35492 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35492 | HTTPS FTP |
-Related structure data
| Related structure data |  8ijqMC  8ijrC  8w9wC  8w9yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35492.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35492.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35492_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35492_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The cryo-EM structure of human sphingomyelin synthase-related pro...
| Entire | Name: The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide |
|---|---|
| Components |
|
-Supramolecule #1: The cryo-EM structure of human sphingomyelin synthase-related pro...
| Supramolecule | Name: The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sphingomyelin synthase-related protein 1
| Macromolecule | Name: Sphingomyelin synthase-related protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number:  sphingomyelin synthase sphingomyelin synthase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.056529 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RLDPEYWKTI LSCIYVFIVF GFTSFIMVIV HERVPDMQTY PPLPDIFLDS VPRIPWAFAM TEVCGMILCY IWLLVLLLHK HRSILLRRL CSLMGTVFLL RCFTMFVTSL SVPGQHLQCT GKIYGSVWEK LHRAFAIWSG FGMTLTGVHT CGDYMFSGHT V VLTMLNFF ...String: RLDPEYWKTI LSCIYVFIVF GFTSFIMVIV HERVPDMQTY PPLPDIFLDS VPRIPWAFAM TEVCGMILCY IWLLVLLLHK HRSILLRRL CSLMGTVFLL RCFTMFVTSL SVPGQHLQCT GKIYGSVWEK LHRAFAIWSG FGMTLTGVHT CGDYMFSGHT V VLTMLNFF VTEYTPRSWN FLHTLSWVLN LFGIFFILAA HEHYSIDVFI AFYITTRLFL YYHTLANTRA YQQSRRARIW FP MFSFFEC NVNGTVPNEY CWPFSKP UniProtKB: Sphingomyelin synthase-related protein 1 |
-Macromolecule #2: N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE
| Macromolecule | Name: N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE type: ligand / ID: 2 / Number of copies: 6 / Formula: 16C |
|---|---|
| Molecular weight | Theoretical: 537.901 Da |
| Chemical component information |  ChemComp-16C: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X


















