-Search query
-Search result
Showing 1 - 50 of 62 items for (author: roux & a)
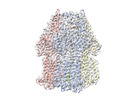
EMDB-17350: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution

PDB-8p1i: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution

EMDB-36794: 
Cryo-EM structure of Na+,K+-ATPase alpha2 from Artemia salina in cation-free E2P form

PDB-8k1l: 
Cryo-EM structure of Na+,K+-ATPase alpha2 from Artemia salina in cation-free E2P form
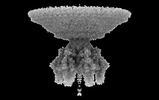
EMDB-41649: 
P22 Mature Virion tail - C6 Localized Reconstruction

EMDB-41651: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution

EMDB-41819: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution

PDB-8tvr: 
In situ cryo-EM structure of bacteriophage P22 tail hub protein: tailspike protein complex at 2.8A resolution

PDB-8tvu: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution

PDB-8u1o: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution

EMDB-41791: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution

EMDB-41792: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution

PDB-8u10: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution

PDB-8u11: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution

EMDB-15802: 
T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA

PDB-8b14: 
T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA

EMDB-10136: 
Double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical membrane bicelle

EMDB-10137: 
Refined, asymmetrically masked double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on helical lipid bicelle

EMDB-10138: 
Segment of helical membrane tube with longitudinal ESCRT-III filaments with different binding modes formed from Snf7/Vps24/Vps2

EMDB-10139: 
Segment of helical membrane tube with longitudinal ESCRT-III filaments in the equatorial binding mode formed from Snf7/Vps24/Vps2

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
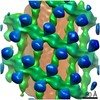
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
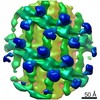
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes

PDB-6rzu: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state

PDB-6rzv: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes

PDB-6rzw: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state

EMDB-9671: 
Adeno-Associated Virus 2 at 2.8 ang

PDB-6ih9: 
Adeno-Associated Virus 2 at 2.8 ang

EMDB-9672: 
Adeno-Associated Virus 2 in complex with AAVR

PDB-6ihb: 
Adeno-Associated Virus 2 in complex with AAVR

EMDB-0007: 
MlaBDEF complex from A. baumannii

PDB-6ic4: 
Cryo-EM structure of the A. baumannii MLA complex at 8.7 A resolution

EMDB-3718: 
3,4-dihydroxybenzoate decarboxylase AroY from Enterobacter cloacae in the apo state

EMDB-6538: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5

EMDB-6539: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5
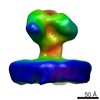
EMDB-6540: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5
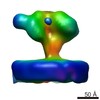
EMDB-6541: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5
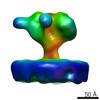
EMDB-6542: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5

EMDB-6543: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5

PDB-3jcb: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5

PDB-3jcc: 
Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5

EMDB-3112: 
Negative stain EM structure of the Tse6-Tsi6-VgrG1-EF-Tu complex in detergent

EMDB-3113: 
Negative stain EM structure of the Tse6-Tsi6-VgrG1-EagT6-EF-Tu complex

EMDB-1813: 
Structural Comparison of HIV-1 Envelope Spikes with and without the V1V2 Loop.

EMDB-1814: 
Structural Comparison of HIV-1 Envelope Spikes with and without the V1V2 Loop

EMDB-1519: 
Cryoelectron tomography of HIV-1 envelope spikes

EMDB-1596: 
HIV-1 Env spike maps using various alignment and classfication combinations
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
