+Search query
-Structure paper
| Title | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery. |
|---|---|
| Journal, issue, pages | J Mol Biol, Vol. 435, Issue 24, Page 168365, Year 2023 |
| Publish date | Dec 15, 2023 |
 Authors Authors | Stephano M Iglesias / Ravi K Lokareddy / Ruoyu Yang / Fenglin Li / Daniel P Yeggoni / Chun-Feng David Hou / Makayla N Leroux / Juliana R Cortines / Justin C Leavitt / Mary Bird / Sherwood R Casjens / Simon White / Carolyn M Teschke / Gino Cingolani /   |
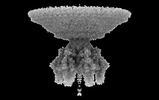
| PubMed Abstract | Bacteriophage P22 is a prototypical member of the Podoviridae superfamily. Since its discovery in 1952, P22 has become a paradigm for phage transduction and a model for icosahedral viral capsid ...Bacteriophage P22 is a prototypical member of the Podoviridae superfamily. Since its discovery in 1952, P22 has become a paradigm for phage transduction and a model for icosahedral viral capsid assembly. Here, we describe the complete architecture of the P22 tail apparatus (gp1, gp4, gp10, gp9, and gp26) and the potential location and organization of P22 ejection proteins (gp7, gp20, and gp16), determined using cryo-EM localized reconstruction, genetic knockouts, and biochemical analysis. We found that the tail apparatus exists in two equivalent conformations, rotated by ∼6° relative to the capsid. Portal protomers make unique contacts with coat subunits in both conformations, explaining the 12:5 symmetry mismatch. The tail assembles around the hexameric tail hub (gp10), which folds into an interrupted β-propeller characterized by an apical insertion domain. The tail hub connects proximally to the dodecameric portal protein and head-to-tail adapter (gp4), distally to the trimeric tail needle (gp26), and laterally to six trimeric tailspikes (gp9) that attach asymmetrically to gp10 insertion domain. Cryo-EM analysis of P22 mutants lacking the ejection proteins gp7 or gp20 and biochemical analysis of purified recombinant proteins suggest that gp7 and gp20 form a molecular complex associated with the tail apparatus via the portal protein barrel. We identified a putative signal transduction pathway from the tailspike to the tail needle, mediated by three flexible loops in the tail hub, that explains how lipopolysaccharide (LPS) is sufficient to trigger the ejection of the P22 DNA in vitro. |
 External links External links |  J Mol Biol / J Mol Biol /  PubMed:37952769 / PubMed:37952769 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 3.4 Å |
| Structure data | EMDB-41649: P22 Mature Virion tail - C6 Localized Reconstruction EMDB-41651, PDB-8tvu: EMDB-41791, PDB-8u10: EMDB-41792, PDB-8u11: EMDB-41819, PDB-8u1o: |
| Source |
|
 Keywords Keywords |  VIRAL PROTEIN / VIRAL PROTEIN /  phage / phage /  bacteriophage / tail spike protein / TSP / gene product 9 (gp9) / Packaged DNA stabilization protein / gene product 10 (gp10) / bacteriophage / tail spike protein / TSP / gene product 9 (gp9) / Packaged DNA stabilization protein / gene product 10 (gp10) /  STRUCTURAL PROTEIN / portal protein / head-to-tail protein / gene product 1 (gp1) / gene product 4 (gp4 / gene product 4 (gp4) / Tail hub protein / gene product (1) / Coat protein / Major capsid protein / gene product 5 (gp5) / Tailspike protein STRUCTURAL PROTEIN / portal protein / head-to-tail protein / gene product 1 (gp1) / gene product 4 (gp4 / gene product 4 (gp4) / Tail hub protein / gene product (1) / Coat protein / Major capsid protein / gene product 5 (gp5) / Tailspike protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers