-Search query
-Search result
Showing 1 - 50 of 96 items for (author: prabu & jr)

EMDB-18214: 
Structure of CUL9-RBX1 ubiquitin E3 ligase complex - hexameric assembly
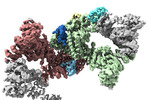
EMDB-18216: 
Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer
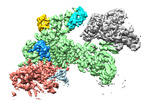
EMDB-18217: 
Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused on E2-like density

EMDB-18218: 
Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused dimeric core
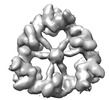
EMDB-18220: 
Structure of the hexameric CUL9-RBX1 complex with deletion of CUL9 CPH domain
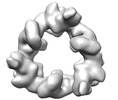
EMDB-18221: 
Structure of the hexameric CUL9-RBX1 complex with deletion of CUL9 DOC domain
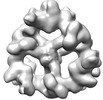
EMDB-18222: 
Structure of the hexameric CUL9-RBX1 complex with deletion of CUL9 ARM9 domain
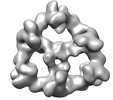
EMDB-18223: 
Structure of the hexameric CUL9-RBX1 complex with deletion of CUL9 ARIH-RBR element

EMDB-19179: 
Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer

EMDB-19856: 
Focused map 1- K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19857: 
Focused map 2 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19858: 
Focused map 3 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19859: 
Focused map 4 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19860: 
Focused map 5 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-18207: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide, Glacios map

EMDB-18230: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide
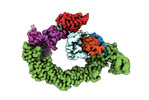
EMDB-18915: 
Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2

EMDB-17798: 
CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1

EMDB-17799: 
CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 2

EMDB-17800: 
NEDD8-CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1

EMDB-17801: 
NEDD8-CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 2

EMDB-17802: 
K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL1-RBX1-SKP1-FBXW7 with trapped UBE2R2-donor UB-acceptor UB-cyclin E peptide

EMDB-17803: 
Consensus map - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide

EMDB-17822: 
K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide

EMDB-18767: 
K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB~acceptor UB-BRD4

EMDB-17705: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class I

EMDB-17706: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class II

EMDB-17707: 
Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class III

EMDB-17709: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class V

EMDB-17710: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class IV

EMDB-17713: 
Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin

EMDB-17715: 
SRS and Cat modules of human CTLHSR4 bound to multiphosphorylated UBE2H~ubiquitin

EMDB-17716: 
Structure of CTLHSR4 - phospho-UBE2H~ubiquitin bound to engineered VH
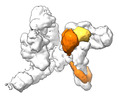
EMDB-17717: 
SRS and Cat modules of yeast Chelator-GIDSR4 bound to multiphosphorylated Ubc8~ubiquitin
EMDB-17764: 
Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin
EMDB-16891: 
Structure of the UBE1L activating enzyme bound to ISG15 and UBE2L6
EMDB-18589: 
Map of UBE1L activating enzyme bound to ISG15 (FL) and UBE2L6

EMDB-16355: 
Structure of Dimeric HECT E3 Ubiquitin Ligase UBR5

EMDB-16356: 
Structure of HECT E3 UBR5 forming K48 linked Ubiquitin chains

EMDB-16865: 
Tetrameric HECT E3 Ubiquitin Ligase UBR5

EMDB-16866: 
Cryo-EM map of ubiquitin-VME bound HECT E3 ligase UBR5

EMDB-16867: 
Ubiquitin transfer from E2 to E3: UBE2D2-ubiquitin linked to HECT E3 ligase UBR5

EMDB-17466: 
Cryo-EM map of HECT E3 ligase UBR5 forming K48 linked ubiquitin chains

EMDB-25026: 
Anaphase Promoting Complex delta APC7

EMDB-25027: 
Anaphase Promoting Complex delta APC7 + UBE2C,substrate,UbV,CDH1

EMDB-12995: 
Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2*

EMDB-12998: 
Structure of Neddylated CRL5Vif-CBFbeta-ARIH2*-APOBEC3C (A3C fullcomplex consensus )

EMDB-12999: 
Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* ( A3G E3-E3 catalytic focused )

EMDB-13000: 
Structure of Neddylated CRL5Vif-CBFbeta-ARIH2*-APOBEC3C (A3C consensus )

EMDB-13001: 
Structure of Neddylated CRL5Vif-CBFbeta-ARIH2*-APOBEC3G ( A3G consensus )
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
