+Search query
-Structure paper
| Title | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 84, Issue 2, Page 293-308.e14, Year 2024 |
| Publish date | Jan 18, 2024 |
 Authors Authors | Jakub Chrustowicz / Dawafuti Sherpa / Jerry Li / Christine R Langlois / Eleftheria C Papadopoulou / D Tung Vu / Laura A Hehl / Özge Karayel / Viola Beier / Susanne von Gronau / Judith Müller / J Rajan Prabu / Matthias Mann / Gary Kleiger / Arno F Alpi / Brenda A Schulman /   |
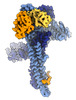
| PubMed Abstract | Ubiquitylation is catalyzed by coordinated actions of E3 and E2 enzymes. Molecular principles governing many important E3-E2 partnerships remain unknown, including those for RING-family GID/CTLH E3 ...Ubiquitylation is catalyzed by coordinated actions of E3 and E2 enzymes. Molecular principles governing many important E3-E2 partnerships remain unknown, including those for RING-family GID/CTLH E3 ubiquitin ligases and their dedicated E2, Ubc8/UBE2H (yeast/human nomenclature). GID/CTLH-Ubc8/UBE2H-mediated ubiquitylation regulates biological processes ranging from yeast metabolic signaling to human development. Here, cryoelectron microscopy (cryo-EM), biochemistry, and cell biology reveal this exquisitely specific E3-E2 pairing through an unconventional catalytic assembly and auxiliary interactions 70-100 Å away, mediated by E2 multisite phosphorylation. Rather than dynamic polyelectrostatic interactions reported for other ubiquitylation complexes, multiple Ubc8/UBE2H phosphorylation sites within acidic CK2-targeted sequences specifically anchor the E2 C termini to E3 basic patches. Positions of phospho-dependent interactions relative to the catalytic domains correlate across evolution. Overall, our data show that phosphorylation-dependent multivalency establishes a specific E3-E2 partnership, is antagonistic with dephosphorylation, rigidifies the catalytic centers within a flexing GID E3-substrate assembly, and facilitates substrate collision with ubiquitylation active sites. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:38113892 / PubMed:38113892 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.4 - 19.5 Å |
| Structure data |  EMDB-17705: Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class I  EMDB-17706: Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class II  EMDB-17707: Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class III  EMDB-17709: Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class V  EMDB-17710: Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class IV EMDB-17713, PDB-8pjn:  EMDB-17715: SRS and Cat modules of human CTLHSR4 bound to multiphosphorylated UBE2H~ubiquitin  EMDB-17716: Structure of CTLHSR4 - phospho-UBE2H~ubiquitin bound to engineered VH  EMDB-17717: SRS and Cat modules of yeast Chelator-GIDSR4 bound to multiphosphorylated Ubc8~ubiquitin EMDB-17764, PDB-8pmq: |
| Chemicals |  ChemComp-ZN: |
| Source |
|
 Keywords Keywords |  LIGASE / LIGASE /  E3 ubiquitin ligase / E3 ubiquitin ligase /  E2 ubiquitin-conjugating enzyme / E2 ubiquitin-conjugating enzyme /  phosphorylation / CTLH / GID / UBE2H phosphorylation / CTLH / GID / UBE2H |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers