-Search query
-Search result
Showing 1 - 50 of 98 items for (author: smith & dy)
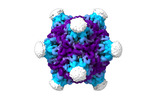
EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
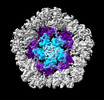
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
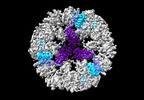
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
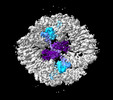
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb3: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb4: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb5: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb7: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcf: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcg: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-27847: 
Mouse norovirus strain CR6, attenuated
Method: single particle / : Smith TJ, Sherman M

EMDB-27849: 
Mouse norovirus strain CR6 at pH 5.0
Method: single particle / : Smith TJ

EMDB-27319: 
Mouse Norovirus strain WU23
Method: single particle / : Smith TJ

EMDB-27321: 
Mouse norovirus strain WU23 + 10mM GCDCA
Method: single particle / : Smith TJ

EMDB-27322: 
Murine norovirus strain WU23 at pH 5
Method: single particle / : Smith TJ

EMDB-31528: 
Ewald sphere-corrected Melbournevirus particle
Method: single particle / : Burton-Smith RN, Murata K

EMDB-31529: 
Fivefold block reconstruction of the Melbournevirus capsid
Method: single particle / : Burton-Smith RN, Murata K

EMDB-31530: 
Threefold block reconstruction of Melbournevirus capsid
Method: single particle / : Burton-Smith RN, Murata K

EMDB-31531: 
Twofold block reconstruction of the Melbournevirus capsid
Method: single particle / : Burton-Smith RN, Murata K

EMDB-25448: 
Negative-stain EM reconstruction of SpFN_1B-06-PL, a SARS-CoV-2 spike fused to H.pylori ferritin nanoparticle vaccine candidate
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25449: 
RFN_131, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Receptor-Binding Domain
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25450: 
pCoV146, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Spike Receptor-Binding and N-Terminal Domains
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25451: 
pCoV111, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Spike S1 Subunit
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-23970: 
Full length SARS-CoV-2 Nsp2
Method: single particle / : QCRG Structural Biology Consortium

EMDB-23971: 
SARS-CoV-2 Nsp2
Method: single particle / : QCRG Structural Biology Consortium

PDB-7msw: 
Full length SARS-CoV-2 Nsp2
Method: single particle / : QCRG Structural Biology Consortium

PDB-7msx: 
SARS-CoV-2 Nsp2
Method: single particle / : QCRG Structural Biology Consortium

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-22295: 
Cryo-EM structure of SHIV-elicited RHA1.V2.01 in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-6xrt: 
Cryo-EM structure of SHIV-elicited RHA1.V2.01 in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

EMDB-10462: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408
Method: single particle / : Srinivas H

EMDB-10463: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib
Method: single particle / : Srinivas H

PDB-6tcz: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408
Method: single particle / : Srinivas H

PDB-6td5: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib
Method: single particle / : Srinivas H

EMDB-10418: 
CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2
Method: single particle / : Wang Q, Pike ACW, Grieben M, Baronina A, Nasrallah C, Shintre C, Edwards AM, Arrowsmith CH, Bountra C, Carpenter EP, Structural Genomics Consortium (SGC)

EMDB-10419: 
CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2
Method: single particle / : Wang Q, Pike ACW, Grieben M, Baronina A, Nasrallah C, Shintre C, Edwards AM, Arrowsmith CH, Bountra C, Carpenter EP, Structural Genomics Consortium (SGC)

PDB-6t9n: 
CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2
Method: single particle / : Wang Q, Pike ACW, Grieben M, Baronina A, Nasrallah C, Shintre C, Edwards AM, Arrowsmith CH, Bountra C, Carpenter EP, Structural Genomics Consortium (SGC)

PDB-6t9o: 
CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2
Method: single particle / : Wang Q, Pike ACW, Grieben M, Baronina A, Nasrallah C, Shintre C, Edwards AM, Arrowsmith CH, Bountra C, Carpenter EP, Structural Genomics Consortium (SGC)

EMDB-20160: 
The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas wild type axoneme
Method: subtomogram averaging / : Fu G, Nicastro D

EMDB-20161: 
The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap92 mutant axoneme
Method: subtomogram averaging / : Fu G, Nicastro D

EMDB-20162: 
The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap76-1 mutant axoneme
Method: subtomogram averaging / : Fu G, Nicastro D

EMDB-20163: 
The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap76-2 mutant axoneme
Method: subtomogram averaging / : Fu G, Nicastro D

EMDB-20164: 
The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap81 mutant axoneme
Method: subtomogram averaging / : Fu G, Nicastro D

EMDB-20165: 
The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap216 mutant axoneme
Method: subtomogram averaging / : Fu G, Nicastro D

EMDB-20166: 
The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap76-1,fap81 double mutant axoneme
Method: subtomogram averaging / : Fu G, Nicastro D

EMDB-0628: 
The axonemal doublet microtubule focusing on the inner junction region extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas wild type cilia
Method: subtomogram averaging / : Lin J, Fu G, Nicastro D, Dymek EE, Porter M, Smith EF

EMDB-0629: 
The axonemal doublet microtubule focusing on the inner junction region extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas pacrg mutant cilia
Method: subtomogram averaging / : Lin J, Fu G, Nicastro D, Dymek EE, Porter M, Smith EF
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
