-Search query
-Search result
Showing 1 - 50 of 139 items for (author: ruiz & t)

EMDB-43514: 
SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface
Method: single particle / : Esfahani BG, Randolph P, Peng R, Grant T, Stroupe ME, Stagg SM

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10
Method: single particle / : Huang J, Ozorowski G, Ward AB

PDB-8sx3: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10
Method: single particle / : Huang J, Ozorowski G, Ward AB

EMDB-19492: 
Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase
Method: single particle / : Ruiz FM, Plaza-Pegueroles A, Fernandez-Tornero C

PDB-8rth: 
Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase
Method: single particle / : Ruiz FM, Plaza-Pegueroles A, Fernandez-Tornero C

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
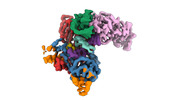
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
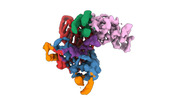
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-16521: 
Cryo-EM structure of the Cibeles-Demetra 3:3 heterocomplex from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E, Araujo-Bazan L, Berger JM

EMDB-16524: 
Cryo-EM structure of the Ceres homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E, Araujo-Bazan L, Berger JM

EMDB-16531: 
Cryo-EM structure of the Cora homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E, Araujo-Bazan L, Berger JM

PDB-8ca9: 
Cryo-EM structure of the Cibeles-Demetra 3:3 heterocomplex from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E

PDB-8cad: 
Cryo-EM structure of the Ceres homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E

PDB-8can: 
Cryo-EM structure of the Cora homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E
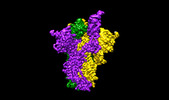
EMDB-17576: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL
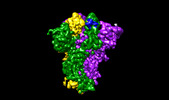
EMDB-17578: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

PDB-8p99: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

PDB-8p9y: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

EMDB-28205: 
Clostridioides difficile binary toxin translocase CDTb wild-type after calcium depletion from receptor binding domain 1 (RBD1) - Class 2
Method: single particle / : Abeyawardhane DL, Pozharski E

EMDB-28206: 
Clostridioides difficile binary toxin translocase CDTb wild-type after calcium depletion from receptor binding domain 1 (RBD1) - Class 1
Method: single particle / : Abeyawardhane DL, Pozharski E

EMDB-28207: 
Clostridioides difficile binary toxin translocase CDTb double mutant - D623A D734A
Method: single particle / : Abeyawardhane DL, Pozharski E

EMDB-16114: 
Vitamin B12 transporter BtuB1 with lipoprotein BtuG1 from B. theta
Method: single particle / : Silale A, Abellon-Ruiz J, van den Berg B

EMDB-17574: 
BtuB3G3 bound to cyanocobalamin with disordered EL8
Method: single particle / : Silale A, Abellon-Ruiz J, van den Berg B

EMDB-17575: 
BtuB3G3 bound to cyanocobalamin with ordered EL8
Method: single particle / : Silale A, Abellon-Ruiz J, van den Berg B

PDB-8blw: 
Vitamin B12 transporter BtuB1 with lipoprotein BtuG1 from B. theta
Method: single particle / : Silale A, Abellon-Ruiz J, van den Berg B

PDB-8p97: 
BtuB3G3 bound to cyanocobalamin with disordered EL8
Method: single particle / : Silale A, Abellon-Ruiz J, van den Berg B

PDB-8p98: 
BtuB3G3 bound to cyanocobalamin with ordered EL8
Method: single particle / : Silale A, Abellon-Ruiz J, van den Berg B

EMDB-15393: 
Cryo-EM structure of a substrate-bound glutamate transporter homologue GltTk encapsulated within a nanodisc
Method: single particle / : Whittaker JJ, Guskov A

PDB-8afa: 
Cryo-EM structure of a substrate-bound glutamate transporter homologue GltTk encapsulated within a nanodisc
Method: single particle / : Whittaker JJ, Guskov A

EMDB-26999: 
CryoEM structure of the T-pilus from Agrobacterium tumefaciens
Method: single particle / : Bui KH, Black CS

EMDB-27023: 
CryoEM structure of the N-pilus from Escherichia coli
Method: single particle / : Bui KH, Black CS

PDB-8cue: 
CryoEM structure of the T-pilus from Agrobacterium tumefaciens
Method: single particle / : Bui KH, Black CS

PDB-8cw4: 
CryoEM structure of the N-pilus from Escherichia coli
Method: single particle / : Bui KH, Black CS

EMDB-27264: 
CryoEM structure of human METTL1-WDR4 in complex with Lys-tRNA
Method: single particle / : Ruiz-Arroyo VM, Nam Y

EMDB-27265: 
CryoEM structure of human METTL1-WDR4 in complex with Lys-tRNA and SAM
Method: single particle / : Ruiz-Arroyo VM, Nam Y

EMDB-28108: 
CryoEM structure of human METTL1-WDR4 in complex with Lys-tRNA and SAH.
Method: single particle / : Ruiz-Arroyo VM, Nam Y

PDB-8d9k: 
CryoEM structure of human METTL1-WDR4 in complex with Lys-tRNA
Method: single particle / : Ruiz-Arroyo VM, Nam Y

PDB-8d9l: 
CryoEM structure of human METTL1-WDR4 in complex with Lys-tRNA and SAM
Method: single particle / : Ruiz-Arroyo VM, Nam Y

PDB-8eg0: 
CryoEM structure of human METTL1-WDR4 in complex with Lys-tRNA and SAH
Method: single particle / : Ruiz-Arroyo VM, Nam Y

EMDB-29052: 
SARS-CoV-2 Spike Hexapro - C68.59 Fab (Class 3 - disordered)
Method: single particle / : Croft JT, Lee KK

EMDB-29053: 
SARS-CoV-2 Spike Hexapro - C59.68 Fab (Class 1 - No Fab bound)
Method: single particle / : Croft JT, Lee KK

EMDB-29054: 
SARS-CoV-2 Spike Hexapro - C68.59 Fab (Class 2 - Fab bound)
Method: single particle / : Croft JT, Lee KK

EMDB-28147: 
Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

EMDB-28259: 
Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

EMDB-28269: 
Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

PDB-8ehg: 
Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

PDB-8emq: 
Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

PDB-8en7: 
Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
