-Search query
-Search result
Showing all 48 items for (author: reyes & fe)

EMDB-16942: 
Murine type II Abeta fibril from APP23 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16944: 
Murine type III Abeta fibril from APP/PS1 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16949: 
Murine type II Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16952: 
Murine type II Abeta fibril from tgAPPSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16953: 
MurineArc type I Abeta fibril from tg-APPArcSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16957: 
DI2 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16959: 
DI1 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16960: 
Murine type III Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16961: 
DI3 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol2: 
Murine type II Abeta fibril from APP23 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol3: 
Murine type III Abeta fibril from APP/PS1 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol5: 
Murine type II Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol6: 
Murine type II Abeta fibril from tgAPPSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol7: 
MurineArc type I Abeta fibril from tg-APPArcSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8olg: 
DI2 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8oln: 
DI1 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8olo: 
Murine type III Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8olq: 
DI3 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-20258: 
Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

PDB-6p5n: 
Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

EMDB-20248: 
Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 1)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

EMDB-20249: 
Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 2)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

EMDB-20255: 
Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 1)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

EMDB-20256: 
Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2)
Method: single particle / : Acosta-Reyes FJ, Neupane R

EMDB-20257: 
Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

PDB-6p4g: 
Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 1)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

PDB-6p4h: 
Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 2)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

PDB-6p5i: 
Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 1)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

PDB-6p5j: 
Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS

PDB-6p5k: 
Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3)
Method: single particle / : Acosta-Reyes FJ, Neupane R, Frank J, Fernandez IS
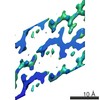
EMDB-8272: 
Human Islet Amyloid Polypeptide Segment 19-SGNNFGAILSS-29 with Early Onset S20G Mutation Determined by MicroED
Method: electron crystallography / : Krotee PAL, Rodriguez JA

EMDB-8273: 
Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED
Method: electron crystallography / : Krotee PAL, Rodriguez JA

EMDB-8196: 
Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED
Method: electron crystallography / : Rodriguez JA, Sawaya MR

EMDB-8197: 
Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED
Method: electron crystallography / : Rodriguez JA, Sawaya MR

EMDB-8198: 
Structure of GNNQQNY from yeast prion Sup35 in space group P21 determined by MicroED
Method: electron crystallography / : Rodriguez JA, Sawaya MR
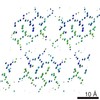
EMDB-8199: 
Structure of GNNQQNY from yeast prion Sup35 in space group P212121 determined by MicroED
Method: electron crystallography / : Rodriguez JA, Sawaya MR

EMDB-8216: 
MicroED structure of tau VQIVYK peptide at 1.1 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8217: 
MicroED structure of lysozyme at 1.8 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8218: 
MicroED structure of xylanase at 2.3 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8219: 
MicroED structure of thaumatin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8220: 
MicroED structure of trypsin at 1.7 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8221: 
MicroED structure of proteinase K at 1.6 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8222: 
MicroED structure of thermolysin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8472: 
MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution
Method: electron crystallography / : Weiss SC, de la Cruz MJ

EMDB-8077: 
MicroED structure of proteinase K at 1.75 A resolution
Method: electron crystallography / : Hattne J, Shi D

EMDB-3001: 
MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein, residues 47-56
Method: electron crystallography / : Rodriguez JA, Ivanova M, Sawaya MR, Cascio D, Reyes F, Shi D, Johnson L, Guenther E, Sangwan S, Hattne J, Nannenga B, Gonen T, Eisenberg D
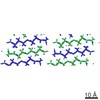
EMDB-3028: 
MicroED structure of the toxic core segment, GAVVTGVTAVA, from Parkinson's disease protein, alpha-synuclein, residues 69-78.
Method: electron crystallography / : Rodriguez JA, Ivanova M, Sawaya MR, Cascio D, Reyes F, Shi D, Johnson L, Guenther E, Zhang M, Jiang L, Arbing MA, Sangwan S, Hattne J, Whitelegge J, Brewster A, Messerschmidt M, Boutet S, Sauter NK, Nannenga B, Gonen T, Eisenberg D
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
