-Search query
-Search result
Showing all 17 items for (author: nieusma & t)

EMDB-20261: 
Single Particle Electron Cryo-Microscopy Reconstruction of BG505 SOSIP-I53-50NP
Method: single particle / : Berndsen ZT, Nieusma T, Ward AB

EMDB-9029: 
Hemagglutinin trimeric ectodomain (B/Massachusetts/02/2012) in complex with Fab from IgG CR9114 and single-domain antibody SD84
Method: single particle / : Pallesen J, Nieusma T, Hoffman RMB, Ward AB
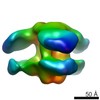
EMDB-8801: 
Negative-stain EM reconstruction of H3v-47 Fab in complex with A/Minnesota/11/2010 H3N2v HA and FI6v Fab
Method: single particle / : Nieusma T, Ward AB

EMDB-8643: 
BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibodies 3BNC117 and PGT145
Method: single particle / : Lee JH, Nieusma T, Ward AB

EMDB-8644: 
BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibody 3BNC117
Method: single particle / : Lee JH, Ward AB

PDB-5v8l: 
BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibodies 3BNC117 and PGT145
Method: single particle / : Lee JH, Ward AB

PDB-5v8m: 
BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibody 3BNC117
Method: single particle / : Lee JH, Cottrell CA, Ward AB

EMDB-3059: 
ZM197 SOSIP.664 trimer in complex with VRC01 Fab
Method: single particle / : Lee JH, Ward AB

EMDB-5834: 
A Unique Human Mycoplasma Protein that Generically Blocks Antigen-Antibody Union
Method: single particle / : Grover RK, Zhu X, Nieusma T, Jones T, Boreo I, MacLeod AS, Mark A, Niessen S, Kim HJ, Kong L, Assad-Garcia N, Kwon K, Chesi M, Salomon DR, Jelinek DF, Kyle RA, Pyles RB, Glass JI, Ward AB, Wilson IA, Lerner RA

EMDB-5835: 
A Unique Human Mycoplasma Protein that Generically Blocks Antigen-Antibody Union
Method: single particle / : Grover RK, Zhu X, Nieusma T, Jones T, Boreo I, MacLeod AS, Mark A, Niessen S, Kim HJ, Kong L, Assad-Garcia N, Kwon K, Chesi M, Salomon DR, Jelinek DF, Kyle RA, Pyles RB, Glass JI, Ward AB, Wilson IA, Lerner RA

EMDB-5836: 
A Unique Human Mycoplasma Protein that Generically Blocks Antigen-Antibody Union
Method: single particle / : Grover RK, Zhu X, Nieusma T, Jones T, Boreo I, MacLeod AS, Mark A, Niessen S, Kim HJ, Kong L, Assad-Garcia N, Kwon K, Chesi M, Salomon DR, Jelinek DF, Kyle RA, Pyles RB, Glass JI, Ward AB, Wilson IA, Lerner RA

EMDB-5759: 
Hepatitis C Virus E2 Envelope Glycoprotein Core Structure
Method: single particle / : Nieusma T, Kong L, Giang E, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Wilson IA, Law M, Ward AB
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
