-Search query
-Search result
Showing 1 - 50 of 354 items for (author: nick & am)

EMDB-41452: 
Cryo-EM structure of monomeric alpha-Klotho
Method: single particle / : Schnicker NJ, Xu Z, Mohammad A, Gakhar L, Huang CL

PDB-8toh: 
Cryo-EM structure of monomeric alpha-Klotho
Method: single particle / : Schnicker NJ, Xu Z, Mohammad A, Gakhar L, Huang CL

EMDB-41256: 
Cryotomogram of DIV 4 cultured hippocampal neuron
Method: electron tomography / : Swulius MT
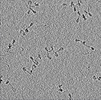
EMDB-41257: 
Purified arrayed human chromatin
Method: electron tomography / : Swulius MT

EMDB-41258: 
In vitro assembled actin and cofilactin filaments
Method: electron tomography / : Swulius MT

EMDB-41263: 
Purified alpha-CaMKII Holoenzymes
Method: electron tomography / : Swulius MT

EMDB-41264: 
DIV 1 hippocampal neuron (weighted back-projection and greyscale segmentation)
Method: electron tomography / : Swulius MT

EMDB-19005: 
structure of the GLMP/MFSD1 complex
Method: single particle / : Jungnickel KEJ, Loew C

EMDB-19006: 
Lysosomal peptide transporter
Method: single particle / : Jungnickel KEJ, Loew C

PDB-8r8q: 
Lysosomal peptide transporter
Method: single particle / : Jungnickel KEJ, Loew C
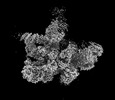
EMDB-18920: 
Plastid-encoded RNA polymerase (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
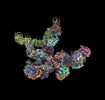
EMDB-18935: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18952: 
Plastid-encoded RNA polymerase transcription elongation complex (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18964: 
Plastid-encoded RNA polymerase (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18965: 
Plastid-encoded RNA polymerase (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18974: 
Plastid-encoded RNA polymerase (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18975: 
Plastid-encoded RNA polymerase (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18976: 
Plastid-encoded RNA polymerase (Region 5)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18982: 
Plastid-encoded RNA polymerase (Region 6)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18983: 
Plastid-encoded RNA polymerase (Region 8)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18985: 
Plastid-encoded RNA polymerase (Region 9)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18986: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18995: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18996: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18998: 
Plastid-encoded RNA polymerase (Region 7)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-19007: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
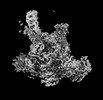
EMDB-19010: 
Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r5o: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r6s: 
Plastid-encoded RNA polymerase (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8rdj: 
Plastid-encoded RNA polymerase transcription elongation complex (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-17111: 
Cryo-EM structure of the wild-type alpha-synuclein fibril.
Method: helical / : Pesch V, Reithofer S, Ma L, Flores-Fernandez JM, Oezduezenciler P, Busch Y, Lien Y, Rudtke O, Frieg B, Schroeder GF, Wille H, Tamgueney G

PDB-8oqi: 
Cryo-EM structure of the wild-type alpha-synuclein fibril.
Method: helical / : Pesch V, Reithofer S, Ma L, Flores-Fernandez JM, Oezduezenciler P, Busch Y, Lien Y, Rudtke O, Frieg B, Schroeder GF, Wille H, Tamgueney G
EMDB-19023: 
Plastid-encoded RNA polymerase transcription elongation complex
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8ras: 
Plastid-encoded RNA polymerase transcription elongation complex
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-29858: 
Hepatitis B virus capsid bound to importin alpha1
Method: single particle / : Yang R, Cingolani G

EMDB-29756: 
Empty capsid of Hepatitis B virus
Method: single particle / : Yang R, Cingolani G

EMDB-29785: 
Hepatitis B virus capsid bound to importin alpha1/beta heterodimer
Method: single particle / : Yang R, Cingolani G

EMDB-42481: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42482: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D

EMDB-42483: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-2heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42485: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42486: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D

PDB-8ur5: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8ur7: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-29452: 
Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor
Method: single particle / : Yu X, Abeywickrema P, Bonneux B, Behera I, Jacoby E, Fung A, Adhikary S, Bhaumik A, Carbajo RJ, Bruyn SD, Miller R, Patrick A, Pham Q, Piassek M, Verheyen N, Shareef A, Sutto-Ortiz P, Ysebaert N, Vlijmen HV, Jonckers THM, Herschke F, McLellan JS, Decroly E, Fearns R, Grosse S, Roymans D, Sharma S, Rigaux P, Jin Z

PDB-8fu3: 
Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor
Method: single particle / : Yu X, Abeywickrema P, Bonneux B, Behera I, Jacoby E, Fung A, Adhikary S, Bhaumik A, Carbajo RJ, Bruyn SD, Miller R, Patrick A, Pham Q, Piassek M, Verheyen N, Shareef A, Sutto-Ortiz P, Ysebaert N, Vlijmen HV, Jonckers THM, Herschke F, McLellan JS, Decroly E, Fearns R, Grosse S, Roymans D, Sharma S, Rigaux P, Jin Z

EMDB-41156: 
HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-41157: 
Global reconstruction for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-41158: 
CS2it1p2_F7K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-41160: 
CS4tt1p1_E3K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
