-Search query
-Search result
Showing all 47 items for (author: mogk & a)

EMDB-18062: 
III2-IV1 respiratory supercomplex from S. pombe
Method: single particle / : Moe A, Brzezinski P

EMDB-18063: 
Local refinement of CIII2 from S. pombe
Method: single particle / : Moe A, Brzezinski P

EMDB-18064: 
Local refinement of CIV from S. pombe
Method: single particle / : Moe A, Brzezinski P

PDB-8q1b: 
III2-IV1 respiratory supercomplex from S. pombe
Method: single particle / : Moe A, Brzezinski P

EMDB-4621: 
Two-Step Activation Mechanism of the ClpB Disaggregase for Sequential Substrate Threading by the Main ATPase Motor.
Method: single particle / : Deville C, Saibil HR

EMDB-4622: 
E. coli ClpB (DWB and K476C mutant) bound to casein - middle domain conformation 1
Method: single particle / : Deville C, Saibil HR

EMDB-4623: 
E. coli ClpB (DWB and K476C mutant) bound to casein - middle domain conformation 2
Method: single particle / : Deville C, Saibil HR

EMDB-4624: 
ClpB (DWB and K476C mutant) bound to casein in presence of ATPgammaS - state KC-1
Method: single particle / : Deville C, Saibil HR

EMDB-4625: 
E. coli ClpB (DWB and K476C mutant) bound to casein - state KC-2
Method: single particle / : Deville C, Saibil HR

EMDB-4626: 
ClpB (DWB and K476C mutant) bound to casein in presence of ATPgammaS - state KC-2A
Method: single particle / : Deville C, Saibil HR

EMDB-4627: 
ClpB (DWB and K476C mutant) bound to casein in presence of ATPgammaS - state KC-2B
Method: single particle / : Deville C, Saibil HR

EMDB-4940: 
ClpB (DWB mutant) bound to casein in presence of ATPgammaS - state WT-1
Method: single particle / : Deville C, Saibil HR

EMDB-4941: 
ClpB (DWB mutant) bound to casein in presence of ATPgammaS - state WT-2A
Method: single particle / : Deville C, Saibil HR

EMDB-4942: 
ClpB (DWB mutant) bound to casein in presence of ATPgammaS - state WT-2B
Method: single particle / : Deville C, Saibil HR

PDB-6qs4: 
Two-Step Activation Mechanism of the ClpB Disaggregase for Sequential Substrate Threading by the Main ATPase Motor.
Method: single particle / : Deville C, Saibil HR

PDB-6qs6: 
ClpB (DWB and K476C mutant) bound to casein in presence of ATPgammaS - state KC-1
Method: single particle / : Deville C, Saibil HR

PDB-6qs7: 
ClpB (DWB and K476C mutant) bound to casein in presence of ATPgammaS - state KC-2A
Method: single particle / : Deville C, Saibil HR

PDB-6qs8: 
ClpB (DWB and K476C mutant) bound to casein in presence of ATPgammaS - state KC-2B
Method: single particle / : Deville C, Saibil HR

PDB-6rn2: 
ClpB (DWB mutant) bound to casein in presence of ATPgammaS - state WT-1
Method: single particle / : Deville C, Saibil HR

PDB-6rn3: 
ClpB (DWB mutant) bound to casein in presence of ATPgammaS - state WT-2A
Method: single particle / : Deville C, Saibil HR

PDB-6rn4: 
ClpB (DWB mutant) bound to casein in presence of ATPgammaS - state WT-2B
Method: single particle / : Deville C, Saibil HR

EMDB-3895: 
S.aureus ClpC resting state, asymmetric map
Method: single particle / : Carroni M, Mogk A, Bukau B, Franke K

EMDB-3897: 
Structure of S.aureus ClpC in complex with MecA
Method: single particle / : Carroni M, Mogk A, Bukau B, Franke K

PDB-6em9: 
S.aureus ClpC resting state, asymmetric map
Method: single particle / : Carroni M, Mogk A, Bukau B, Franke K

PDB-6emw: 
Structure of S.aureus ClpC in complex with MecA
Method: single particle / : Carroni M, Mogk A, Bukau B, Franke K

EMDB-3776: 
Cryo EM structure of the bacterial disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein
Method: single particle / : Deville C, Carroni M, Franke KB, Topf M, Bukau B, Mogk A, Saibil HR

EMDB-3777: 
Cryo EM structure of the bacterial disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state
Method: single particle / : Deville C, Carroni M, Franke KB, Topf M, Bukau B, Mogk A, Saibil HR

PDB-5ofo: 
Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein
Method: single particle / : Deville C, Carroni M, Franke KB, Topf M, Bukau B, Mogk A, Saibil HR

PDB-5og1: 
Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state
Method: single particle / : Deville C, Carroni M, Franke KB, Topf M, Bukau B, Mogk A, Saibil HR

EMDB-3094: 
Electron negative stain tomography of alpha-synuclein amyloid fibrils
Method: electron tomography / : Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B

EMDB-3095: 
Electron negative stain tomography of alpha-synuclein amyloid fibrils treated with the Hsc70/DNAJB1/Apg2 chaperone system
Method: electron tomography / : Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B

EMDB-2524: 
Cryo EM structure of the contractile Type Six Secretion System VipA/B complex
Method: helical / : Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P

EMDB-2525: 
Cryo EM structure of the contractile Type Six Secretion System VipA/B complex
Method: helical / : Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P

PDB-4d2q: 
Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR

PDB-4d2u: 
Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR

PDB-4d2x: 
Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR

EMDB-2555: 
Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR

EMDB-2556: 
Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR

EMDB-2557: 
Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR

EMDB-2558: 
Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR
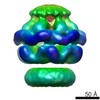
EMDB-2559: 
Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR
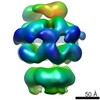
EMDB-2560: 
Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR

EMDB-2561: 
Negative-stain electron microscopy of Hsp104 (HAP form bound to ClpP)
Method: single particle / : Carroni M, Kummer E, Oguchi Y, Clare DK, Wendler P, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
