[English] 日本語
 Yorodumi
Yorodumi- EMDB-4940: ClpB (DWB mutant) bound to casein in presence of ATPgammaS - stat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4940 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | ClpB (DWB mutant) bound to casein in presence of ATPgammaS - state WT-1 | ||||||||||||
 Map data Map data | main cryoEM map of ClpB-DWB casein in presence of ATPgS -state WT-1 | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to heat / protein refolding /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / ATP binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Bos taurus (cattle) / Bos taurus (cattle) /   Escherichia coli HVH 50 (4-2593475) (bacteria) Escherichia coli HVH 50 (4-2593475) (bacteria) | ||||||||||||
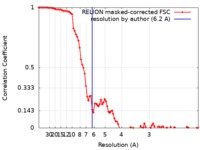
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.2 Å cryo EM / Resolution: 6.2 Å | ||||||||||||
 Authors Authors | Deville C / Saibil HR | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: Two-Step Activation Mechanism of the ClpB Disaggregase for Sequential Substrate Threading by the Main ATPase Motor. Authors: Célia Deville / Kamila Franke / Axel Mogk / Bernd Bukau / Helen R Saibil /   Abstract: AAA+ proteins form asymmetric hexameric rings that hydrolyze ATP and thread substrate proteins through a central channel via mobile substrate-binding pore loops. Understanding how ATPase and ...AAA+ proteins form asymmetric hexameric rings that hydrolyze ATP and thread substrate proteins through a central channel via mobile substrate-binding pore loops. Understanding how ATPase and threading activities are regulated and intertwined is key to understanding the AAA+ protein mechanism. We studied the disaggregase ClpB, which contains tandem ATPase domains (AAA1, AAA2) and shifts between low and high ATPase and threading activities. Coiled-coil M-domains repress ClpB activity by encircling the AAA1 ring. Here, we determine the mechanism of ClpB activation by comparing ATPase mechanisms and cryo-EM structures of ClpB wild-type and a constitutively active ClpB M-domain mutant. We show that ClpB activation reduces ATPase cooperativity and induces a sequential mode of ATP hydrolysis in the AAA2 ring, the main ATPase motor. AAA1 and AAA2 rings do not work synchronously but in alternating cycles. This ensures high grip, enabling substrate threading via a processive, rope-climbing mechanism. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4940.map.gz emd_4940.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4940-v30.xml emd-4940-v30.xml emd-4940.xml emd-4940.xml | 28.3 KB 28.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4940_fsc.xml emd_4940_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4940.png emd_4940.png | 68 KB | ||
| Masks |  emd_4940_msk_1.map emd_4940_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_4940_additional_1.map.gz emd_4940_additional_1.map.gz emd_4940_additional_2.map.gz emd_4940_additional_2.map.gz emd_4940_half_map_1.map.gz emd_4940_half_map_1.map.gz emd_4940_half_map_2.map.gz emd_4940_half_map_2.map.gz | 49.5 MB 40.4 MB 49.8 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4940 http://ftp.pdbj.org/pub/emdb/structures/EMD-4940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4940 | HTTPS FTP |
-Related structure data
| Related structure data |  6rn2MC  4621C  4622C  4623C  4624C  4625C  4626C  4627C  4941C  4942C  6qs4C  6qs6C  6qs7C  6qs8C  6rn3C  6rn4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4940.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4940.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main cryoEM map of ClpB-DWB casein in presence of ATPgS -state WT-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4940_msk_1.map emd_4940_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
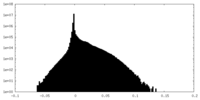
| Density Histograms |
-Additional map: non postprocessed cryoEM map of ClpB-DWB casein in...
| File | emd_4940_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non postprocessed cryoEM map of ClpB-DWB casein in presence of ATPgS -state WT-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
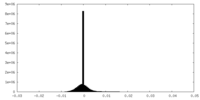
| Density Histograms |
-Additional map: cryoEM map filtered according to local resolution of...
| File | emd_4940_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM map filtered according to local resolution of ClpB-DWB casein in presence of ATPgS -state WT-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
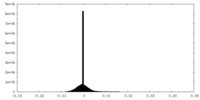
| Density Histograms |
-Half map: odd cryoEM half map of ClpB-DWB casein in...
| File | emd_4940_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | odd cryoEM half map of ClpB-DWB casein in presence of ATPgS -state WT-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: even cryoEM half map of ClpB-DWB casein in...
| File | emd_4940_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | even cryoEM half map of ClpB-DWB casein in presence of ATPgS -state WT-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
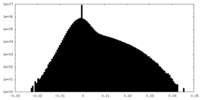
| Density Histograms |
- Sample components
Sample components
-Entire : ClpB-DWB bound to casein in presence of ATPgammaS
| Entire | Name: ClpB-DWB bound to casein in presence of ATPgammaS |
|---|---|
| Components |
|
-Supramolecule #1: ClpB-DWB bound to casein in presence of ATPgammaS
| Supramolecule | Name: ClpB-DWB bound to casein in presence of ATPgammaS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Supramolecule #2: ClpB-DWB
| Supramolecule | Name: ClpB-DWB / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Supramolecule #3: casein
| Supramolecule | Name: casein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
-Macromolecule #1: Chaperone protein ClpB
| Macromolecule | Name: Chaperone protein ClpB / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli HVH 50 (4-2593475) (bacteria) / Strain: 4-2593475 Escherichia coli HVH 50 (4-2593475) (bacteria) / Strain: 4-2593475 |
| Molecular weight | Theoretical: 95.734172 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MRLDRLTNKF QLALADAQSL ALGHDNQFIE PLHLMSALLN QEGGSVSPLL TSAGINAGQL RTDINQALNR LPQVEGTGGD VQPSQDLVR VLNLCDKLAQ KRGDNFISSE LFVLAALESR GTLADILKAA GATTANITQA IEQMRGGESV NDQGAEDQRQ A LKKYTIDL ...String: MRLDRLTNKF QLALADAQSL ALGHDNQFIE PLHLMSALLN QEGGSVSPLL TSAGINAGQL RTDINQALNR LPQVEGTGGD VQPSQDLVR VLNLCDKLAQ KRGDNFISSE LFVLAALESR GTLADILKAA GATTANITQA IEQMRGGESV NDQGAEDQRQ A LKKYTIDL TERAEQGKLD PVIGRDEEIR RTIQVLQRRT KNNPVLIGEP GVGKTAIVEG LAQRIINGEV PEGLKGRRVL AL DMGALVA GAKYRGEFEE RLKGVLNDLA KQEGNVILFI DALHTMVGAG KADGAMDAGN MLKPALARGE LHCVGATTLD EYR QYIEKD AALERRFQKV FVAEPSVEDT IAILRGLKER YELHHHVQIT DPAIVAAATL SHRYIADRQL PDKAIDLIDE AASS IRMQI DSKPEELDRL DRRIIQLKLE QQALMKESDE ASKKRLDMLN EELSDKERQY SELEEEWKAE KASLSGTQTI KAELE QAKI AIEQARRVGD LARMSELQYG KIPELEKQLE AATQLEGKTM RLLRNKVTDA EIAEVLARWT GIPVSRMMES EREKLL RME QELHHRVIGQ NEAVDAVSNA IRRSRAGLAD PNRPIGSFLF LGPTGVGKTE LCKALANFMF DSDEAMVRID MSEFMEK HS VSRLVGAPPG YVGYEEGGYL TEAVRRRPYS VILLDAVEKA HPDVFNILLQ VLDDGRLTDG QGRTVDFRNT VVIMTSNL G SDLIQERFGE LDYAHMKELV LGVVSHNFRP EFINRIDEVV VFHPLGEQHI ASIAQIQLKR LYKRLEERGY EIHISDEAL KLLSENGYDP VYGARPLKRA IQQQIENPLA QQILSGELVP GKVIRLEVNE DRIVAVQH |
-Macromolecule #2: casein
| Macromolecule | Name: casein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 1.723883 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: AAAAAAAAAA AAAAAAAAAA AAAA |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 9 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.6 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1687 / Average electron dose: 1.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X