-Search query
-Search result
Showing 1 - 50 of 155 items for (author: lowe & j)

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcf: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcg: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-15395: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15398: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2, symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15399: 
Cryo-EM map of crescentin filament in complex with a megabody (stutter mutant, C2 symmetry, large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15400: 
Cryo-EM map of crescentin filaments in complex with a megabody (wildtype, C2, large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15401: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15402: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

EMDB-15446: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15465: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15473: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

EMDB-15476: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8afe: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8afh: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2, symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8afl: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8afm: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and small box)
Method: single particle / : Liu Y, Lowe J

PDB-8ahl: 
Cryo-EM structure of crescentin filaments (stutter mutant, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8aia: 
Cryo-EM structure of crescentin filaments (wildtype, C1 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8aix: 
Cryo-EM structure of crescentin filaments (wildtype, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J

PDB-8ajb: 
Cryo-EM structure of crescentin filaments (stutter mutant, C2 symmetry and large box)
Method: single particle / : Liu Y, Lowe J
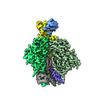
EMDB-29248: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
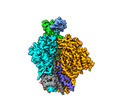
EMDB-29264: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
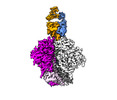
EMDB-29288: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fk5: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fl1: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8flw: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

EMDB-16042: 
Core divisome complex FtsWIQBL from Pseudomonas aeruginosa
Method: single particle / : Kaeshammer L, van den Ent F, Jeffery M, Lowe J

PDB-8bh1: 
Core divisome complex FtsWIQBL from Pseudomonas aeruginosa
Method: single particle / : Kaeshammer L, van den Ent F, Jeffery M, Lowe J

EMDB-15205: 
cryoEM structure of the catalytically inactive EndoS from S. pyogenes in complex with the Fc region of immunoglobulin G1
Method: single particle / : Trastoy B, Cifuente JO, Du JJ, Sundberg EJ, Guerin ME

PDB-8a64: 
cryoEM structure of the catalytically inactive EndoS from S. pyogenes in complex with the Fc region of immunoglobulin G1.
Method: single particle / : Trastoy B, Cifuente JO, Du JJ, Sundberg EJ, Guerin ME

EMDB-14776: 
Signal peptide mimicry primes Sec61 for client-selective inhibition
Method: single particle / : Rehan S, Paavilainen O V

PDB-7zl3: 
Signal peptide mimicry primes Sec61 for client-selective inhibition
Method: single particle / : Rehan S, Paavilainen O V

EMDB-29323: 
Structure of RdrA from Escherichia coli RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

EMDB-29324: 
Map of RdrA from Escherichia coli RADAR defense system in single-split conformation
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

EMDB-29325: 
Map of RdrA from Escherichia coli RADAR defense system in double-split conformation
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

EMDB-29326: 
Structure of RdrA from Streptococcus suis RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

EMDB-29327: 
Structure of RdrB from Escherichia coli RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

EMDB-29328: 
Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

PDB-8fnt: 
Structure of RdrA from Escherichia coli RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

PDB-8fnu: 
Structure of RdrA from Streptococcus suis RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

PDB-8fnv: 
Structure of RdrB from Escherichia coli RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

PDB-8fnw: 
Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system
Method: single particle / : Duncan-Lowey B, Johnson AG, Rawson S, Mayer ML, Kranzusch PJ

EMDB-14150: 
D. melanogaster 13-protofilament microtubule
Method: single particle / : Wagstaff J, Planelles-Herrero VJ, Derivery E, Lowe J

PDB-7qup: 
D. melanogaster 13-protofilament microtubule
Method: single particle / : Wagstaff J, Planelles-Herrero VJ, Derivery E, Lowe J

EMDB-14147: 
D. melanogaster alpha/beta tubulin heterodimer in the GDP form
Method: single particle / : Wagstaff J, Planelles-Herrero VJ, Derivery E, Lowe J

EMDB-14148: 
D. melanogaster alpha/beta tubulin heterodimer in the GTP form
Method: single particle / : Wagstaff J, Planelles-Herrero VJ, Derivery E, Lowe J

EMDB-14151: 
BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament (Prosthecobacter dejongeii)
Method: single particle / : Wagstaff J, Planelles-Herrero VJ

PDB-7quc: 
D. melanogaster alpha/beta tubulin heterodimer in the GDP form
Method: single particle / : Wagstaff J, Planelles-Herrero VJ, Derivery E, Lowe J
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
