-Search query
-Search result
Showing 1 - 50 of 57 items for (author: lee & jw)

EMDB-36937: 
post-occluded structure of human ABCB6 W546A mutant (ADP/VO4-bound)
Method: single particle / : Jin MS, Lee SS, Park JG, Jang E, Choi SH, Kim S, Kim JW

EMDB-36938: 
Outward-facing structure of human ABCB6 W546A mutant (ADP/VO4-bound)
Method: single particle / : Jin MS, Lee SS, Park JG, Jang E, Choi SH, Kim S, Kim JW

EMDB-33734: 
Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody
Method: single particle / : Yoo Y, Cho HS

EMDB-28915: 
SIRT6 bound to an H3K9Ac nucleosome
Method: single particle / : Markert J, Whedon S, Wang Z, Cole P, Farnung L

EMDB-28551: 
RMC-5552 in complex with mTORC1 and FKBP12
Method: single particle / : Tomlinson ACA, Yano JK

EMDB-32844: 
Toll-like receptor3 linear cluster
Method: single particle / : Lim CS, Jang YH, Lee GY, Han GM, Lee JO

EMDB-32845: 
ectoTLR3-poly(I:C) cluster
Method: single particle / : Lim CS, Jang YH, Lee GY, Han GM, Lee JO

EMDB-32851: 
CT-mut (D523K,D524K,E527K) TLR3-poly(I:C) complex
Method: single particle / : Lim CS, Jang YH, Lee GY, Han GM, Lee JO

EMDB-32852: 
ectoTLR3-mAb12-poly(I:C) complex
Method: single particle / : Lim CS, Jang YH, Lee GY, Han GM, Lee JO

EMDB-32853: 
NT-mut(K117D,K139D,K145D) TLR3 -poly I:C complex
Method: single particle / : Lim CS, Jang YH, Lee GY, Han GM, Lee JO

EMDB-34361: 
TLR3(A795H)-poly(I:C) complex
Method: single particle / : Lim CS, Jang YH, Lee GY, Han GM, Lee JO

EMDB-27735: 
Cryo-EM structure of SIVmac239 SOS-2P Env trimer in complex with human bNAb PGT145
Method: single particle / : Gorman J, Kwong PD

EMDB-27718: 
SIV E660.CR54 SOS-2P Env Trimer with ITS92.02
Method: single particle / : Gorman J, Kwong PD

EMDB-25062: 
In-situ structure of SIV trimer
Method: subtomogram averaging / : Gorman J, Wang CY, Mason RD, Nazzari AF, Welles H, Zhou TQ, Jr JB, Tsybovsky Y, Verardi R, Yang YP, Zhang BS, Lifson JD, Liu J, Roederer M, Kwong PD

EMDB-25063: 
in situ SIVmac239-Env trimers on the surface of AT-2-inactivated virions
Method: subtomogram averaging / : Liu J, Wang CY

EMDB-25064: 
in situ SIVmac239-Env trimers on the surface of AT-2-inactivated virions
Method: subtomogram averaging / : Liu J, Wang CY

EMDB-25065: 
In-situ structure of SIVmac239-Env trimers with ITS90.03 associated
Method: subtomogram averaging / : Liu J, Wang CY

EMDB-27631: 
SIV mac239 SOS-2P K169T Env trimer with bNAb PGT145 and jacalin
Method: single particle / : Gorman J, Kwong PD

EMDB-30790: 
Cryo-EM structure of the human ABCB6 (coproporphyrin III-bound)
Method: single particle / : Kim S, Lee SS, Park JG, Kim JW, Kim NJ, Hong S, Kang JY, Jin MS

EMDB-30791: 
Cryo-EM structure of the human ABCB6 (Hemin and GSH-bound)
Method: single particle / : Kim S, Lee SS, Park JG, Kim JW, Kim NJ, Hong S, Kang JY, Jin MS

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-0103: 
Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses
Method: single particle / : Geraets JA, Flatt JW, Domanska A, Butcher SJ
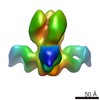
EMDB-5793: 
A common solution to group 2 influenza virus neutralization
Method: single particle / : Friesen RHE, Leeb PS, Stoop EJM, Hoffman RMB, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, Korse HJWM, Ophorst C, Brinkman-van der Linden ECM, Throsby M, Kwakkenbos MJ, Bakkerd AQ, Beaumont Y, Spits H, Kwaks T, Vogels R, Ward AB, Goudsmit J, Wilson IA
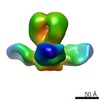
EMDB-5794: 
A common solution to group 2 influenza virus neutralization
Method: single particle / : Friesen RHE, Leeb PS, Stoop EJM, Hoffman RMB, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, Korse HJWM, Ophorst C, Brinkman-van der Linden ECM, Throsby M, Kwakkenbos MJ, Bakkerd AQ, Beaumont Y, Spits H, Kwaks T, Vogels R, Ward AB, Goudsmit J, Wilson IA

EMDB-2248: 
negative stain single-particle reconstruction of conformation I of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
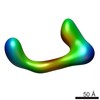
EMDB-2249: 
negative stain single-particle reconstruction of conformation II of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2250: 
negative stain single-particle reconstruction of conformation III of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
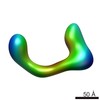
EMDB-2251: 
negative stain single-particle reconstruction of conformation IV of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
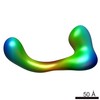
EMDB-2252: 
negative stain single-particle reconstruction of conformation V of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
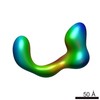
EMDB-2253: 
negative stain single-particle reconstruction of conformation VI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
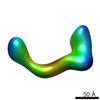
EMDB-2254: 
negative stain single-particle reconstruction of conformation VII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
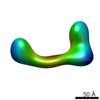
EMDB-2255: 
negative stain single-particle reconstruction of conformation VIII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2256: 
negative stain single-particle reconstruction of conformation IX of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
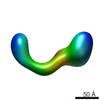
EMDB-2257: 
negative stain single-particle reconstruction of conformation X of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2258: 
negative stain single-particle reconstruction of conformation XI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2259: 
negative stain single-particle reconstruction of conformation XII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2260: 
negative stain single-particle reconstruction of conformation XIII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2261: 
negative stain single-particle reconstruction of conformation XIV of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2262: 
negative stain single-particle reconstruction of conformation XV of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2263: 
negative stain single-particle reconstruction of conformation XVI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
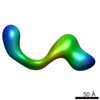
EMDB-2264: 
negative stain single-particle reconstruction of conformation XVII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2265: 
negative stain single-particle reconstruction of conformation XVIII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2266: 
negative stain single-particle reconstruction of conformation XIX of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2267: 
negative stain single-particle reconstruction of conformation XX of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2268: 
negative stain single-particle reconstruction of conformation XXI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
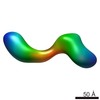
EMDB-2269: 
negative stain single-particle reconstruction of conformation XXII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2143: 
Highly Conserved Protective Epitopes on Influenza B Viruses
Method: single particle / : Khayat R, Lee JH, Dreyfus C, Laursen NS, Wilson IA, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
