-Search query
-Search result
Showing 1 - 50 of 51 items for (author: eisenberg & de)

EMDB-41171: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, wild-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L

EMDB-41172: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, variant-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L
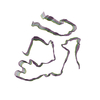
PDB-8tdn: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, wild-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L
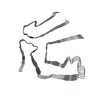
PDB-8tdo: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, variant-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L

PDB-8e7e: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient
Method: helical / : Nguyen BA, Singh V, Afrin S, Saelices L

PDB-8e7j: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient
Method: helical / : Nguyen BA, Saelices L

EMDB-27323: 
Cardiac amyloid fibrils extracted from a variant ATTR I84S amyloidosis patient (2).
Method: helical / : Nguyen BA, Saelices L

EMDB-26685: 
Cardiac amyloid fibrils extracted from a variant ATTR I84S amyloidosis patient
Method: helical / : Nguyen BA, Saelices L

EMDB-26663: 
Tau Paired Helical Filament from Alzheimer's Disease not incubated with EGCG
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS

EMDB-26664: 
Tau Paired Helical Filament from Alzheimer's Disease incubated 1 hr. with EGCG
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS

EMDB-26665: 
Tau Paired Helical Filament from Alzheimer's Disease incubated with EGCG for 3 hours
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS
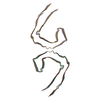
PDB-7upe: 
Tau Paired Helical Filament from Alzheimer's Disease not incubated with EGCG
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS
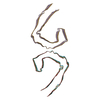
PDB-7upf: 
Tau Paired Helical Filament from Alzheimer's Disease incubated 1 hr. with EGCG
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS
PDB-7upg: 
Tau Paired Helical Filament from Alzheimer's Disease incubated with EGCG for 3 hours
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS

EMDB-24953: 
Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 1
Method: helical / : Cao Q, Jiang Y, Sawaya MR, Eisenberg DS

EMDB-24954: 
Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2
Method: helical / : Cao Q, Jiang Y, Sawaya MR, Eisenberg DS

EMDB-24955: 
Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 3
Method: helical / : Cao Q, Jiang Y, Sawaya MR, Eisenberg DS

PDB-7saq: 
Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 1
Method: helical / : Cao Q, Jiang Y, Sawaya MR, Eisenberg DS

PDB-7sar: 
Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2
Method: helical / : Cao Q, Jiang Y, Sawaya MR, Eisenberg DS

PDB-7sas: 
Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 3
Method: helical / : Cao Q, Jiang Y, Sawaya MR, Eisenberg DS

EMDB-23686: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 1
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

EMDB-23687: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 2
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

EMDB-23688: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 3
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

EMDB-23689: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 4
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

PDB-7m61: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 1
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS
PDB-7m62: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 2
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

PDB-7m64: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 3
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

PDB-7m65: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 4
Method: helical / : Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

PDB-6nk4: 
KVQIINKKL, crystal structure of a tau protein fragment
Method: electron crystallography / : Eisenberg DS, Boyer DR, Sawaya MR

EMDB-7017: 
Segment from bank vole prion protein 168-176 QYNNQNNFV
Method: electron crystallography / : Glynn C, Rodriguez JA

EMDB-7287: 
MicroED structure of carbamazepine at 0.85 A resolution
Method: electron crystallography / : Gallagher-Jones M, Glynn C, Boyer DR, Martynowycz MW, Hernandez E, Miao J, Zee CT, NoviKova IV, Goldschmidt L, McFarlane HT, Helguera GF, Evans JE, Sawaya MR, Cascio D, Eisenberg D, Gonen T, Rodriguez JA

PDB-6axz: 
Segment from bank vole prion protein 168-176 QYNNQNNFV
Method: electron crystallography / : Glynn C, Rodriguez JA, Boyer DR, Gallagher-Jones M
PDB-5ty4: 
MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution
Method: electron crystallography / : Weiss SC, de la Cruz MJ, Hattne J, Shi D, Reyes FE, Callero G, Gonen T

PDB-5k7n: 
MicroED structure of tau VQIVYK peptide at 1.1 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7o: 
MicroED structure of lysozyme at 1.8 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7p: 
MicroED structure of xylanase at 2.3 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7q: 
MicroED structure of thaumatin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7r: 
MicroED structure of trypsin at 1.7 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7s: 
MicroED structure of proteinase K at 1.6 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7t: 
MicroED structure of thermolysin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8216: 
MicroED structure of tau VQIVYK peptide at 1.1 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8217: 
MicroED structure of lysozyme at 1.8 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8218: 
MicroED structure of xylanase at 2.3 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8219: 
MicroED structure of thaumatin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8220: 
MicroED structure of trypsin at 1.7 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8221: 
MicroED structure of proteinase K at 1.6 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8222: 
MicroED structure of thermolysin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8472: 
MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution
Method: electron crystallography / : Weiss SC, de la Cruz MJ
EMDB-2379: 
Structure of herpesvirus fusion glycoprotein B-bilayer complex revealing the protein-membrane and lateral protein-protein interaction
Method: subtomogram averaging / : Maurer UE, Zeev-Ben-Mordehai Z, Pandurangan AP, Cairns TM, Hannah BP, Whitbeck JC, Eisenberg RJ, Cohen GH, Topf M, Huiskonen JT, Grunewald K

EMDB-2380: 
Structure of herpesvirus fusion glycoprotein B-bilayer complex revealing the protein-membrane and lateral protein-protein interaction
Method: subtomogram averaging / : Maurer UE, Zeev-Ben-Mordehai Z, Pandurangan AP, Cairns TM, Hannah BP, Whitbeck JC, Eisenberg RJ, Cohen GH, Topf M, Huiskonen JT, Grunewald K
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
