-Search query
-Search result
Showing all 49 items for (author: dai & jb)

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D
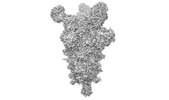
EMDB-26676: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26677: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26678: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-27043: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody SP1-77 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K
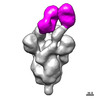
EMDB-27044: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-7-53 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

EMDB-27046: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

EMDB-22378: 
Herpes Simplex Virus Type 1 Procapsid with Portal Vertex
Method: subtomogram averaging / : Buch MHC, Heymann JB, Newcomb WW, Winkler DC, Steven AC

EMDB-22379: 
Herpes Simplex Virus Type 1 A-capsid with Portal Vertex
Method: subtomogram averaging / : Buch MHC, Heymann JB, Newcomb WW, Winkler DC, Steven AC

EMDB-22925: 
cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, two RBDs bound
Method: single particle / : Li Z, Rini JM

EMDB-22926: 
cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, three RBDs bound
Method: single particle / : Li Z, Rini JM

EMDB-23064: 
SARS-CoV-2 spike protein in complex with Fab 15033-7, 3-"up", asymmetric
Method: single particle / : Li Z, Rini J

EMDB-23065: 
SARS-CoV-2 spike protein in complex with Fab 15033-7, 2-"up"-1-"down" conformation
Method: single particle / : Li Z, Rini J

EMDB-22331: 
SPN3US phage empty capsid
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-22332: 
Phage SPN3US mottled capsid (glutaraldehyde-fixed)
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-22333: 
SPN3US phage mottled capsid
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-22078: 
Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis
Method: single particle / : Herrera NG, Morano NC, Celikgil A, Georgiev GI, Malonis R, Lee JH, Tong K, Vergnolle O, Massimi A, Yen LY, Noble AJ, Kopylov M, Bonanno JB, Garrett-Thompson SC, Hayes DB, Brenowitz M, Garforth SJ, Eng ET, Lai JR, Almo SC

EMDB-7907: 
Two retinoschisin molecules interacting laterally forming a unit cell of a filamentous strand.
Method: single particle / : Heymann JB, Vijayasarathy C, Huang RK, Dearborn AD, Sieving PA, Steven AC

EMDB-8607: 
Herpes Simplex Virion Primary Enveloped Virion
Method: single particle / : Fontana J, Newcomb WW, Winkler DC, Cheng N, Heymann JB, Steven AC

EMDB-6439: 
Helical cryo-EM reconstruction of HIV Rev filament
Method: helical / : DiMattia MA, Watts NR, Cheng N, Huang R, Heymann JB, Wingfield PT, Grimes JM, Stuart DI, Steven AC

EMDB-3367: 
Cryo-EM structure of yeast cytoplasmic exosome
Method: single particle / : Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao WW, Zhou K, Liu QS, Dai JB, Yang XR, Dong MQ, Huang N, Wang HW

EMDB-3368: 
Cryo-EM structure of yeast cytoplasmic exosome
Method: single particle / : Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao WW, Zhou K, Liu QS, Dai JB, Yang XR, Dong MQ, Huang N, Wang HW

EMDB-3369: 
Cryo-EM structure of yeast cytoplasmic exosome
Method: single particle / : Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao WW, Zhou K, Liu QS, Dai JB, Yang XR, Dong MQ, Huang N, Wang HW

EMDB-3370: 
Cryo-EM structure of yeast cytoplasmic exosome
Method: single particle / : Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao WW, Zhou K, Liu QS, Dai JB, Yang XR, Dong MQ, Huang N, Wang HW

EMDB-3371: 
Cryo-EM structure of yeast cytoplasmic exosome
Method: single particle / : Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao WW, Zhou K, Liu QS, Dai JB, Yang XR, Dong MQ, Huang N, Wang HW

EMDB-3372: 
Cryo-EM structure of yeast cytoplasmic exosome
Method: single particle / : Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao WW, Zhou K, Liu QS, Dai JB, Yang XR, Dong MQ, Huang N, Wang HW

EMDB-3366: 
Cryo-EM structure of yeast cytoplasmic exosome
Method: single particle / : Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao WW, Zhou K, Liu QS, Dai JB, Yang XR, Dong MQ, Huang N, Wang HW

EMDB-6425: 
Retinoschisin, back-to-back octameric rings
Method: single particle / : Heymann JB, Tolun G, Vijayasarathy C, Huang R, Zeng Y, Li Y, Steven AC, Sieving PA

EMDB-6482: 
Cryo-electron microscopy of alpha Synuclein amyloid fibrils
Method: helical / : Dearborn AD, Wall JS, Cheng N, Heymann JB, Kajava AV, Varkey J, Langen R, Steven AC

EMDB-5917: 
Encapsulin protein (EncA) from Myxococcus xanthus
Method: single particle / : McHugh CA, Fontana J, Lam AS, Cheng N, Aksyuk AA, Steven AC, Hoiczyk E

EMDB-5953: 
Cryo-electron microscopy of iron-core lacking encapsulin nanocompartments from Myxococcus xanthus
Method: single particle / : McHugh CA, Fontana J, Nemecek D, Cheng N, Aksyuk AA, Heymann JB, Winkler DC, Lam AS, Wall JS, Steven AC, Hoiczyk E

EMDB-5772: 
A Two-Pronged Structural Analysis of Retroviral Maturation Indicates that Core Formation Proceeds by a Disassembly-Reassembly Pathway Rather than a Displacive Transition
Method: single particle / : Keller PW, Huang RK, England M, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC

EMDB-5773: 
A Two-Pronged Structural Analysis of Retroviral Maturation Indicates that Core Formation Proceeds by a Disassembly-Reassembly Pathway Rather than a Displacive Transition
Method: single particle / : Keller PW, Huang RK, England M, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC

EMDB-5774: 
A Two-Pronged Structural Analysis of Retroviral Maturation Indicates that Core Formation Proceeds by a Disassembly-Reassembly Pathway Rather than a Displacive Transition
Method: single particle / : Keller PW, Huang RK, England M, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC

EMDB-2364: 
CryoEM reconstruction of the bacteriophage phi6 procapsid to the near-atomic resolution
Method: single particle / : Nemecek D, Boura E, Wu W, Cheng N, Plevka P, Qiao J, Mindich L, Heymann JB, Hurley JH, Steven AC

EMDB-2341: 
Bacteriophage phi6 procapsids with different composition of accessory proteins
Method: single particle / : Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-2342: 
Bacteriophage phi6 procapsids with different composition of accessory proteins
Method: single particle / : Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-2344: 
Bacteriophage phi6 procapsids with different composition of accessory proteins
Method: single particle / : Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-2346: 
Bacteriophage phi6 procapsids with different composition of accessory proteins
Method: single particle / : Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-5355: 
Expansion Intermediates of the Bacteriophage phi6 Procapsid
Method: single particle / : Nemecek D, Cheng N, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-5356: 
Expansion Intermediates of the Bacteriophage phi6 Procapsid
Method: single particle / : Nemecek D, Cheng N, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-5357: 
Expansion Intermediates of the Bacteriophage phi6 Procapsid
Method: single particle / : Nemecek D, Cheng N, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-5358: 
RNA-packaged Capsid of Bacteriophage phi6
Method: single particle / : Nemecek D, Cheng N, Qiao J, Mindich L, Steven AC, Heymann JB

EMDB-1500: 
Initial location of the RNA-dependent RNA polymerase in the bacteriophage phi6 procapsid determined by cryo-electron microscopy
Method: single particle / : Sen A, Heymann JB, Cheng N, Qiao J, Mindich L, Steven AC

EMDB-1501: 
Initial location of the RNA-dependent RNA polymerase in the bacteriophage phi6 procapsid determined by cryo-electron microscopy
Method: single particle / : Sen A, Heymann JB, Cheng N, Qiao J, Mindich L, Steven AC
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
