-Search query
-Search result
Showing 1 - 50 of 61 items for (author: christensen & n)

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
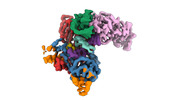
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
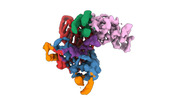
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
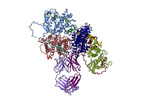
EMDB-14438: 
VAR2 complex with PAM1.4
Method: single particle / : Raghavan SSR, Wang KT

EMDB-14446: 
VAR2CSA APO
Method: single particle / : Raghavan SSR, Wang KT

PDB-7z12: 
VAR2 complex with PAM1.4
Method: single particle / : Raghavan SSR, Wang KT

PDB-7z1h: 
VAR2CSA APO
Method: single particle / : Raghavan SSR, Wang KT

EMDB-24741: 
Cryo EM analysis reveals inherent flexibility of authentic murine papillomavirus capsids
Method: single particle / : Hartmann SR, Hafenstein S

PDB-7ryj: 
Cryo EM analysis reveals inherent flexibility of authentic murine papillomavirus capsids
Method: single particle / : Hartmann SR, Hafenstein S

EMDB-11978: 
Nanobody E bound to Spike-RBD in a localized reconstruction
Method: single particle / : Hallberg BM, Das H

EMDB-11981: 
SARS-CoV-spike bound to two neutralising nanobodies
Method: single particle / : Hallberg BM, Das H

PDB-7b14: 
Nanobody E bound to Spike-RBD in a localized reconstruction
Method: single particle / : Hallberg BM, Das H

PDB-7b18: 
SARS-CoV-spike bound to two neutralising nanobodies
Method: single particle / : Hallberg BM, Das H

EMDB-23081: 
High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility
Method: single particle / : Hartmann SR, Goetschius DJ

PDB-7kzf: 
High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility
Method: single particle / : Hartmann SR, Goetschius DJ, Hafenstein S

EMDB-11980: 
SARS-CoV-spike RBD bound to two neutralising nanobodies.
Method: single particle / : Hallberg BM, Das H

PDB-7b17: 
SARS-CoV-spike RBD bound to two neutralising nanobodies.
Method: single particle / : Hallberg BM, Das H

EMDB-23018: 
SARS-CoV-2 spike in complex with nanobodies E
Method: single particle / : Hallberg BM, Das H

PDB-7ksg: 
SARS-CoV-2 spike in complex with nanobodies E
Method: single particle / : Hallberg BM, Das H

EMDB-22640: 
Murine polyomavirus pentavalent capsomer with 8A7H5 Fab, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22641: 
Murine polyomavirus hexavalent capsomer with 8A7H5 Fab, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22642: 
Murine polyomavirus pentavalent capsomer, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22643: 
Murine polyomavirus hexavalent capsomer, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

PDB-7k22: 
Murine polyomavirus pentavalent capsomer with 8A7H5 Fab, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

PDB-7k23: 
Murine polyomavirus hexavalent capsomer with 8A7H5 Fab, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

PDB-7k24: 
Murine polyomavirus pentavalent capsomer, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

PDB-7k25: 
Murine polyomavirus hexavalent capsomer, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22645: 
Murine polyomavirus (icosahedral reconstruction)
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22646: 
Murine polyomavirus with 8A7H5 Fab (icosahedral reconstruction)
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-20562: 
HIV-1 CA hexamer from TRIM5alpha-coated tubes
Method: subtomogram averaging / : Skorupka KA, Pornillos O, Ganser-Pornillos BK

EMDB-20563: 
TRIM5alpha dimer in complex with HIV-1 capsid tubes
Method: electron tomography / : Skorupka KA, Pornillos O, Ganser-Pornillos BK

EMDB-20564: 
TRIM5alpha trimer bound to HIV-1 capsid tubes
Method: subtomogram averaging / : Skorupka KA, Pornillos O, Ganser-Pornillos BK

EMDB-20565: 
TRIM5alpha lattice bound to HIV-1 capsid lattice
Method: subtomogram averaging / : Skorupka KA, Pornillos O, Ganser-Pornillos BK

EMDB-20574: 
TRIM5alpha-coated HIV-1 capsid tube
Method: electron tomography / : Skorupka KA, Pornillos O, Ganser-Pornillos BK

EMDB-7136: 
High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitope on Human Papillomavirus 16
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christenson ND, Hafenstein S

PDB-6bsp: 
High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitope on Human Papillomavirus 16
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christenson ND, Hafenstein S

PDB-6bt3: 
High-Resolution Structure Analysis of Antibody V5 Conformational Epitope on Human Papillomavirus 16
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-8243: 
Near-atomic resolution cryo-EM reconstruction of HPV16 complexed with Fab V5
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-6619: 
Electron cryo-microscopy of Human Papillomavirus
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-6620: 
Electron cryo-microscopy of Human Papillomavirus
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

PDB-5kep: 
High resolution cryo-EM maps of Human Papillomavirus 16 reveal L2 location and heparin-induced conformational changes
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

PDB-5keq: 
High resolution cryo-EM maps of Human papillomavirus 16 reveal L2 location and heparin-induced conformational changes
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-6423: 
The U4 antibody epitope on human papillomavirus 16 identified by cryo-EM
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Christensen ND, Hafenstein S

EMDB-6424: 
The U4 antibody epitope on human papillomavirus 16 identified by cryo-EM
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Christensen ND, Hafenstein S

PDB-3jba: 
The U4 antibody epitope on human papillomavirus 16 identified by cryo-EM
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Christensen ND, Hafenstein S

EMDB-5990: 
Electron cryo-microscopy of quasi-human papillomavirus 16 complexed with Fab H16.1A
Method: single particle / : Guan J, Brendle S, Bywaters S, Lee H, Ashley RE, Conway JF, Makhov AM, Christensen N, Hafenstein S

EMDB-6121: 
Cryo-EM reconstruction of quasi-HPV16 complexed with H16.14J Fab
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Conway JF, Makhov AM, Christensen ND, Hafenstein S

EMDB-6184: 
Cryo-EM reconstruction of quasi-HPV16 complexed with H263.A2 Fab
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Lee H, Ashley R, Conway JF, Makhov AM, Christensen ND, Hafenstein S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
