+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9uaj | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ovorubin from the golden apple snail (Pomacea canaliculata) | ||||||||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | LIPID BINDING PROTEIN / ovorubin / golden apple snail / Pomacea canaliculata | ||||||||||||||||||||||||||||||
| Function / homology | (Z)-docos-13-enoic acid / Uncharacterized protein / Perivitellin protein / Perivitellin ovorubin-2 / Perivitellin ovorubin-1 Function and homology information Function and homology information | ||||||||||||||||||||||||||||||
| Biological species |  Pomacea canaliculata (invertebrata) Pomacea canaliculata (invertebrata) | ||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.12 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Wangkanont, K. / Saw, W.-G. / Tran, B.N. / Wilasluck, P. | ||||||||||||||||||||||||||||||
| Funding support |  Thailand, 1items Thailand, 1items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Protein Sci / Year: 2026 Journal: Protein Sci / Year: 2026Title: Structure of the chromoprotein ovorubin from the golden apple snail (Pomacea canaliculata). Authors: Patcharin Wilasluck / Wuan-Geok Saw / Bich Ngoc Tran / Grzegorz Sabat / Orion Shih / Kowit Hengphasatporn / Yasuteru Shigeta / Nawaporn Vinayavekhin / Kittikhun Wangkanont /      Abstract: The golden apple snail (Pomacea canaliculata), an invasive gastropod, produces distinctly bright pink egg masses. The astaxanthin-binding ovorubin (P. canaliculata ovorubin [PcOvo]) is a 300-kDa ...The golden apple snail (Pomacea canaliculata), an invasive gastropod, produces distinctly bright pink egg masses. The astaxanthin-binding ovorubin (P. canaliculata ovorubin [PcOvo]) is a 300-kDa glycoprotein responsible for the egg coloration. Here, we determine the three-dimensional structure of PcOvo using cryo-electron microscopy (cryo-EM). PcOvo is a heterodecameric protein consisting of two copies of each PcOvo1-5 subunit. The subunits have similar repeated ferredoxin-like structures. N-linked glycosylation sites are identified. Solution x-ray scattering data support the overall architecture of the complex. PcOvo3 and PcOvo5 have hydrophobic pockets that likely bind various hydrophobic compounds. The cryo-EM map and binding experiments suggest that PcOvo5 binds astaxanthin, which is responsible for the pink color of the protein. Our structure provides molecular insights into the nature of the gastropod egg coloration and lays a foundation for further investigation of ovorubin biology and evolution. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9uaj.cif.gz 9uaj.cif.gz | 385.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9uaj.ent.gz pdb9uaj.ent.gz | 309.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9uaj.json.gz 9uaj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ua/9uaj https://data.pdbj.org/pub/pdb/validation_reports/ua/9uaj ftp://data.pdbj.org/pub/pdb/validation_reports/ua/9uaj ftp://data.pdbj.org/pub/pdb/validation_reports/ua/9uaj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  63984MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 3 types, 6 molecules AFBJDH
| #1: Protein | Mass: 22841.211 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Pomacea canaliculata (invertebrata) / References: UniProt: A0A2T7NVP5 Pomacea canaliculata (invertebrata) / References: UniProt: A0A2T7NVP5#2: Protein | Mass: 22252.473 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Pomacea canaliculata (invertebrata) / References: UniProt: A0A2T7NVP5 Pomacea canaliculata (invertebrata) / References: UniProt: A0A2T7NVP5#4: Protein | Mass: 22467.189 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Pomacea canaliculata (invertebrata) / References: UniProt: A0A2T7NVP6 Pomacea canaliculata (invertebrata) / References: UniProt: A0A2T7NVP6 |
|---|
-Perivitellin ovorubin- ... , 2 types, 4 molecules CIEG
| #3: Protein | Mass: 22408.697 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Pomacea canaliculata (invertebrata) / References: UniProt: J7I2T6 Pomacea canaliculata (invertebrata) / References: UniProt: J7I2T6#5: Protein | Mass: 23827.533 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Pomacea canaliculata (invertebrata) / References: UniProt: J7HZ90 Pomacea canaliculata (invertebrata) / References: UniProt: J7HZ90 |
|---|
-Sugars , 3 types, 12 molecules 
| #6: Polysaccharide | alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta- ...alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source #7: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source #9: Sugar | |
|---|
-Non-polymers , 2 types, 323 molecules 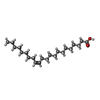


| #8: Chemical | ChemComp-08O / ( #10: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Ovorubin / Type: COMPLEX / Entity ID: #1-#5 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 0.226 MDa / Experimental value: YES |
| Source (natural) | Organism:  Pomacea canaliculata (invertebrata) Pomacea canaliculata (invertebrata) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.5 |
| Buffer component | Conc.: 50 mM / Name: Tris |
| Specimen | Conc.: 9.9 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: COPPER / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.5 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 215000 X / Nominal defocus max: 1400 nm / Nominal defocus min: 400 nm / Cs: 2.7 mm / C2 aperture diameter: 50 µm / Alignment procedure: ZEMLIN TABLEAU |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 2 sec. / Electron dose: 40 e/Å2 / Detector mode: COUNTING / Film or detector model: FEI FALCON IV (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 6418 |
| EM imaging optics | Energyfilter name: TFS Selectris X / Energyfilter slit width: 10 eV |
| Image scans | Width: 4096 / Height: 4096 |
- Processing
Processing
| EM software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1621962 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.12 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 336633 / Algorithm: FOURIER SPACE / Num. of class averages: 5 / Symmetry type: POINT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | B value: 51.4 / Protocol: FLEXIBLE FIT / Space: REAL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Source name: Other / Type: other
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj

