[English] 日本語
 Yorodumi
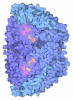
Yorodumi- PDB-9qr3: Methyl-coenzyme M reductase of an ANME-2c from a microbial enrichment -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9qr3 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Methyl-coenzyme M reductase of an ANME-2c from a microbial enrichment | ||||||||||||||||||
 Components Components |
| ||||||||||||||||||
 Keywords Keywords | TRANSFERASE / methanotrophy / anaerobic methanotrophic archaea / methane oxidation / ANME / methyl-coenzyme M reductase / MCR / reverse methanogenesis / nickel-dependent enzyme / coenzyme M / coenzyme B / F430 cofactor | ||||||||||||||||||
| Function / homology | 1-THIOETHANESULFONIC ACID / FACTOR 430 / : / Coenzyme B Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  Candidatus Methanogasteraceae archaeon (archaea) Candidatus Methanogasteraceae archaeon (archaea) | ||||||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.34 Å MOLECULAR REPLACEMENT / Resolution: 1.34 Å | ||||||||||||||||||
 Authors Authors | Mueller, M.-C. / Wagner, T. | ||||||||||||||||||
| Funding support | European Union,  Germany, Germany,  Netherlands, 5items Netherlands, 5items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Atomic resolution structures of the methane-activating enzyme in anaerobic methanotrophy reveal extensive post-translational modifications. Authors: Muller, M.C. / Wissink, M. / Mukherjee, P. / Von Possel, N. / Laso-Perez, R. / Engilberge, S. / Carpentier, P. / Kahnt, J. / Wegener, G. / Welte, C.U. / Wagner, T. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9qr3.cif.gz 9qr3.cif.gz | 5.7 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9qr3.ent.gz pdb9qr3.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  9qr3.json.gz 9qr3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qr/9qr3 https://data.pdbj.org/pub/pdb/validation_reports/qr/9qr3 ftp://data.pdbj.org/pub/pdb/validation_reports/qr/9qr3 ftp://data.pdbj.org/pub/pdb/validation_reports/qr/9qr3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  9qm5C  9qqtC  9qr1C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 3 types, 24 molecules ADGJMPSVBEHKNQTWCFILORUX
| #1: Protein | Mass: 61795.473 Da / Num. of mol.: 8 / Source method: isolated from a natural source Details: The alpha subunit contains seven post-translational modifications: Methylhistidine: 266 Methylarginine: 280 Methylglutamine: 410 Hydroxytryptophane: 437 Thioglycine: 455 Didehydroaspartate: ...Details: The alpha subunit contains seven post-translational modifications: Methylhistidine: 266 Methylarginine: 280 Methylglutamine: 410 Hydroxytryptophane: 437 Thioglycine: 455 Didehydroaspartate: 460 Methylcysteine: 462 Source: (natural)  Candidatus Methanogasteraceae archaeon (archaea) Candidatus Methanogasteraceae archaeon (archaea)Cell line: / / Organ: / / Plasmid details: / / Variant: / / Tissue: / / References: coenzyme-B sulfoethylthiotransferase #2: Protein | Mass: 46238.676 Da / Num. of mol.: 8 / Source method: isolated from a natural source Source: (natural)  Candidatus Methanogasteraceae archaeon (archaea) Candidatus Methanogasteraceae archaeon (archaea)Cell line: / / Organ: / / Plasmid details: / / Variant: / / Tissue: / / References: coenzyme-B sulfoethylthiotransferase #3: Protein | Mass: 29393.064 Da / Num. of mol.: 8 / Source method: isolated from a natural source Source: (natural)  Candidatus Methanogasteraceae archaeon (archaea) Candidatus Methanogasteraceae archaeon (archaea)References: coenzyme-B sulfoethylthiotransferase |
|---|
-Non-polymers , 10 types, 9908 molecules 


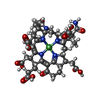















| #4: Chemical | ChemComp-K / #5: Chemical | ChemComp-NA / #6: Chemical | ChemComp-CL / #7: Chemical | ChemComp-F43 / #8: Chemical | ChemComp-COM / #9: Chemical | ChemComp-TP7 / #10: Chemical | ChemComp-GOL / #11: Chemical | ChemComp-MPD / ( #12: Chemical | ChemComp-EDO / #13: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.42 Å3/Da / Density % sol: 49.29 % / Description: Yellow rectangles/squares |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: 2 ul protein at 24.6 g/l in 25 mM Tris/HCl pH 7.6, 10 % v/v glycerol, and 2 mM dithiothreitol was mixed with 2 ul crystallisation solution (30 % v/v 2-Methyl-2,4-pentanediol, 100 mM Tris pH ...Details: 2 ul protein at 24.6 g/l in 25 mM Tris/HCl pH 7.6, 10 % v/v glycerol, and 2 mM dithiothreitol was mixed with 2 ul crystallisation solution (30 % v/v 2-Methyl-2,4-pentanediol, 100 mM Tris pH 8.5, 500 mM NaCl, and 8% polyethylene glycol 8000) on a Junior Clover plate (Jena Bioscience). The reservoir contained 90 ul of crystallisation solution. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.97856 Å / Beamline: PROXIMA 1 / Wavelength: 0.97856 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Apr 28, 2022 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97856 Å / Relative weight: 1 |
| Reflection | Resolution: 1.34→127.16 Å / Num. obs: 1527343 / % possible obs: 96.2 % / Redundancy: 6.3 % / CC1/2: 0.991 / Rmerge(I) obs: 0.168 / Rpim(I) all: 0.073 / Rrim(I) all: 0.183 / Net I/σ(I): 6.6 |
| Reflection shell | Resolution: 1.34→1.5 Å / Redundancy: 6 % / Rmerge(I) obs: 1.671 / Mean I/σ(I) obs: 1.6 / Num. unique obs: 76367 / CC1/2: 0.56 / Rpim(I) all: 0.737 / Rrim(I) all: 1.829 / % possible all: 66.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.34→98.93 Å / SU ML: 0.1 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 16.45 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.34→98.93 Å / SU ML: 0.1 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 16.45 / Stereochemistry target values: MLDetails: The model was built and corrected with COOT (Version 0.8.9.2) and refined with PHENIX.refine. All atoms were considered anisotropic, and hydrogens were added in riding mode. The model was ...Details: The model was built and corrected with COOT (Version 0.8.9.2) and refined with PHENIX.refine. All atoms were considered anisotropic, and hydrogens were added in riding mode. The model was validated by the MolProbity server (http://molprobity.biochem.duke.edu).
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.77 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.34→98.93 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj