[English] 日本語
 Yorodumi
Yorodumi- PDB-9o0u: Crystal structure of CRAF/MEK1 complex with PLX4720 and CH5126766 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9o0u | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of CRAF/MEK1 complex with PLX4720 and CH5126766 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE / CRAF-MEK1 complex / MAPK pathway | ||||||
| Function / homology |  Function and homology information Function and homology informationepithelial cell proliferation involved in lung morphogenesis / death-inducing signaling complex assembly / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / intermediate filament cytoskeleton organization / regulation of axon regeneration / mitogen-activated protein kinase kinase / positive regulation of muscle contraction ...epithelial cell proliferation involved in lung morphogenesis / death-inducing signaling complex assembly / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / intermediate filament cytoskeleton organization / regulation of axon regeneration / mitogen-activated protein kinase kinase / positive regulation of muscle contraction / Golgi inheritance / placenta blood vessel development / MAP-kinase scaffold activity / cerebellar cortex formation / labyrinthine layer development / regulation of Rho protein signal transduction / melanosome transport / type B pancreatic cell proliferation / Signaling by MAP2K mutants / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / Rap1 signalling / vesicle transport along microtubule / positive regulation of axonogenesis / positive regulation of Ras protein signal transduction / insulin secretion involved in cellular response to glucose stimulus / regulation of Golgi inheritance / mitogen-activated protein kinase kinase kinase binding / central nervous system neuron differentiation / trachea formation / triglyceride homeostasis / Negative feedback regulation of MAPK pathway / regulation of early endosome to late endosome transport / IFNG signaling activates MAPKs / regulation of stress-activated MAPK cascade / Frs2-mediated activation / GP1b-IX-V activation signalling / MAPK3 (ERK1) activation / ERBB2-ERBB3 signaling pathway / neurotrophin TRK receptor signaling pathway / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / face development / endodermal cell differentiation / pseudopodium / MAP kinase kinase activity / Bergmann glial cell differentiation / positive regulation of ATP biosynthetic process / regulation of cell differentiation / thyroid gland development / Uptake and function of anthrax toxins / positive regulation of protein serine/threonine kinase activity / extrinsic apoptotic signaling pathway via death domain receptors / somatic stem cell population maintenance / positive regulation of peptidyl-serine phosphorylation / MAP kinase kinase kinase activity / protein kinase activator activity / type II interferon-mediated signaling pathway / Schwann cell development / response to axon injury / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of protein-containing complex assembly / keratinocyte differentiation / neuron projection morphogenesis / response to muscle stretch / ERK1 and ERK2 cascade / myelination / positive regulation of autophagy / protein serine/threonine/tyrosine kinase activity / CD209 (DC-SIGN) signaling / dendrite cytoplasm / insulin-like growth factor receptor signaling pathway / response to glucocorticoid / thymus development / MAP3K8 (TPL2)-dependent MAPK1/3 activation / adenylate cyclase activator activity / protein serine/threonine kinase activator activity / Signal transduction by L1 / cell motility / wound healing / positive regulation of transcription elongation by RNA polymerase II / RAF activation / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation / small GTPase binding / Stimuli-sensing channels / chemotaxis / neuron differentiation / Negative regulation of MAPK pathway / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / cellular senescence / Signaling by BRAF and RAF1 fusions / insulin receptor signaling pathway / late endosome / MAPK cascade / heart development / response to oxidative stress / protein tyrosine kinase activity Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.91 Å MOLECULAR REPLACEMENT / Resolution: 2.91 Å | ||||||
 Authors Authors | Jang, D.M. / Eck, M.J. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2025 Journal: J.Biol.Chem. / Year: 2025Title: Characterization and inhibitor sensitivity of ARAF, BRAF, and CRAF kinases. Authors: Tkacik, E. / Jang, D.M. / Boxer, K. / Ha, B.H. / Eck, M.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9o0u.cif.gz 9o0u.cif.gz | 512.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9o0u.ent.gz pdb9o0u.ent.gz | 385.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9o0u.json.gz 9o0u.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/o0/9o0u https://data.pdbj.org/pub/pdb/validation_reports/o0/9o0u ftp://data.pdbj.org/pub/pdb/validation_reports/o0/9o0u ftp://data.pdbj.org/pub/pdb/validation_reports/o0/9o0u | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  9o0vC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| 4 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 8 molecules ACEGBDFH
| #1: Protein | Mass: 32005.803 Da / Num. of mol.: 4 / Mutation: Y340D, Y341D Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: RAF1, RAF / Production host: Homo sapiens (human) / Gene: RAF1, RAF / Production host:  References: UniProt: P04049, non-specific serine/threonine protein kinase #2: Protein | Mass: 43576.039 Da / Num. of mol.: 4 / Mutation: S218A, S222A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MAP2K1, MEK1, PRKMK1 / Production host: Homo sapiens (human) / Gene: MAP2K1, MEK1, PRKMK1 / Production host:  References: UniProt: Q02750, mitogen-activated protein kinase kinase |
|---|
-Non-polymers , 4 types, 53 molecules 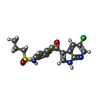






| #3: Chemical | ChemComp-324 / #4: Chemical | ChemComp-CHU / #5: Chemical | ChemComp-ANP / | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.87 Å3/Da / Density % sol: 57.14 % |
|---|---|
| Crystal grow | Temperature: 295 K / Method: vapor diffusion, hanging drop Details: 18% PEG 3350, 0.1 M Sodium citrate pH 5.6, 4% Tacsimate pH 5 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS-II NSLS-II  / Beamline: 17-ID-1 / Wavelength: 0.9202 Å / Beamline: 17-ID-1 / Wavelength: 0.9202 Å |
| Detector | Type: DECTRIS EIGER X 9M / Detector: PIXEL / Date: Nov 8, 2024 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9202 Å / Relative weight: 1 |
| Reflection | Resolution: 2.91→50 Å / Num. obs: 147338 / % possible obs: 99.7 % / Redundancy: 4.79 % / Biso Wilson estimate: 67.3 Å2 / CC1/2: 0.995 / Rmerge(I) obs: 0.132 / Net I/σ(I): 9.63 |
| Reflection shell | Resolution: 2.91→3.08 Å / Redundancy: 4.69 % / Rmerge(I) obs: 1.044 / Num. unique obs: 23539 / CC1/2: 0.995 / % possible all: 98.8 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.91→46.1 Å / SU ML: 0.3576 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 25.7681 MOLECULAR REPLACEMENT / Resolution: 2.91→46.1 Å / SU ML: 0.3576 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 25.7681 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 80.51 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.91→46.1 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj






















