| 機能・相同性 |  機能・相同性情報 機能・相同性情報
: / host cell membrane / viral genome replication / methyltransferase activity / endonuclease activity / symbiont-mediated degradation of host mRNA / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / methylation / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity ...: / host cell membrane / viral genome replication / methyltransferase activity / endonuclease activity / symbiont-mediated degradation of host mRNA / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / methylation / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / single-stranded RNA binding / host cell perinuclear region of cytoplasm / viral protein processing / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont-mediated activation of host autophagy / cysteine-type endopeptidase activity / proteolysis / zinc ion binding / membrane類似検索 - 分子機能 Non-structural protein NSP3, SUD-N (Mac2) domain, betacoronavirus / Sarbecovirus Nsp3c-N domain profile. / Non-structural protein NSP3, N-terminal, betacoronavirus / Polyprotein cleavage domain PL2pro superfamily, betacoronavirus / Non-structural protein NSP3, SUD-N (Mac2) domain superfamily, betacoronavirus / Betacoronavirus SUD-C domain / Betacoronavirus replicase NSP3, N-terminal / NSP1 globular domain superfamily, betacoronavirus / Non-structural protein 2, SARS-CoV-like / Betacoronavirus Nsp3c-M domain profile. ...Non-structural protein NSP3, SUD-N (Mac2) domain, betacoronavirus / Sarbecovirus Nsp3c-N domain profile. / Non-structural protein NSP3, N-terminal, betacoronavirus / Polyprotein cleavage domain PL2pro superfamily, betacoronavirus / Non-structural protein NSP3, SUD-N (Mac2) domain superfamily, betacoronavirus / Betacoronavirus SUD-C domain / Betacoronavirus replicase NSP3, N-terminal / NSP1 globular domain superfamily, betacoronavirus / Non-structural protein 2, SARS-CoV-like / Betacoronavirus Nsp3c-M domain profile. / NSP1, globular domain, betacoronavirus / Non-structural protein NSP3, SUD-M domain, betacoronavirus / Non-structural protein NSP3, SUD-M domain superfamily, betacoronavirus / Betacoronavirus replicase NSP1 / Betacoronavirus single-stranded poly(A) binding domain / NSP1, C-terminal domain, betacoronavirus / : / Betacoronavirus (BetaCoV) Nsp1 C-terminal domain profile. / Betacoronavirus Nsp3e group 2-specific marker (G2M) domain profile. / Betacoronavirus Nsp3c-C domain profile. / Betacoronavirus Nsp3e nucleic acid-binding (NAB) domain profile. / DPUP/SUD, C-terminal, betacoronavirus / Non-structural protein NSP3, nucleic acid-binding domain, betacoronavirus / Non-structural protein NSP3A domain-like superfamily / Non-structural protein NSP3, nucleic acid-binding domain superfamily, betacoronavirus / Non-structural protein 6, betacoronavirus / Betacoronavirus nucleic acid-binding (NAB) / Papain-like viral protease, palm and finger domains, coronavirus / Papain-like protease, N-terminal domain superfamily, coronavirus / Coronavirus replicase NSP2, N-terminal / : / Coronavirus replicase NSP2, C-terminal / Coronavirus (CoV) Nsp2 middle domain profile. / NSP1, globular domain, alpha/betacoronavirus / Coronavirus (CoV) Nsp1 globular domain profile. / Coronavirus (CoV) Nsp2 N-terminal domain profile. / Coronavirus (CoV) Nsp2 C-terminal domain profile. / Nonstructural protein 2, N-terminal domain, coronavirus / Non-structural protein 2, C-terminal domain, coronavirus / NSP3, second ubiquitin-like (Ubl) domain, coronavirus / Coronavirus Nsp3a Ubl domain profile. / Coronavirus Nsp3d Ubl domain profile. / NSP3, first ubiquitin-like (Ubl) domain, coronavirus / Peptidase family C16 domain profile. / : / : / Coronavirus replicase NSP7 / Coronavirus 3Ecto domain profile. / Coronavirus RNA-dependent RNA polymerase (RdRp) Nsp7 cofactor domain profile. / Coronavirus RNA-dependent RNA polymerase (RdRp) Nsp8 cofactor domain profile. / Coronavirus Nsp9 single-stranded RNA (ssRNA)-binding domain profile. / Coronavirus (CoV) ExoN/MTase coactivator domain profile. / Coronavirus (CoV) Nsp3 Y domain profile. / Coronavirus Nsp4 C-terminal (Nsp4C) domain profile. / Coronavirus main protease (M-pro) domain profile. / Peptidase C30, coronavirus / Peptidase C16, coronavirus / Non-structural protein NSP9, coronavirus / Non-structural protein NSP7, coronavirus / Non-structural protein NSP8, coronavirus / RNA synthesis protein NSP10, coronavirus / Non-structural protein NSP4, C-terminal, coronavirus / RNA synthesis protein NSP10 superfamily, coronavirus / Non-structural protein NSP9 superfamily, coronavirus / Non-structural protein NSP7 superfamily, coronavirus / Non-structural protein NSP8 superfamily, coronavirus / Non-structural protein NSP4, C-terminal superfamily, coronavirus / Papain-like protease, thumb domain superfamily, coronavirus / Peptidase C30, domain 3, coronavirus / Non-structural protein 6, coronavirus / Coronavirus replicase NSP3, C-terminal / Non-structural protein NSP4, N-terminal, coronavirus / Coronavirus endopeptidase C30 / Coronavirus papain-like peptidase / Coronavirus replicase NSP8 / Coronavirus RNA synthesis protein NSP10 / Coronavirus replicase NSP4, C-terminal / Coronavirus replicase NSP6 / Coronavirus replicase NSP4, N-terminal / Coronavirus replicase NSP3, C-terminal / Coronavirus replicase NSP9 / Non-structural protein 3, X-domain-like / Appr-1"-p processing enzyme / Macro domain / Macro domain profile. / Macro domain / Macro domain-like / Peptidase S1, PA clan, chymotrypsin-like fold / Peptidase S1, PA clan類似検索 - ドメイン・相同性 ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide / Chem-FHR / ORF1a polyprotein類似検索 - 構成要素 |
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.13 Å
分子置換 / 解像度: 2.13 Å  データ登録者
データ登録者 引用
引用 ジャーナル: Commun Biol / 年: 2025
ジャーナル: Commun Biol / 年: 2025 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 9lug.cif.gz
9lug.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb9lug.ent.gz
pdb9lug.ent.gz PDB形式
PDB形式 9lug.json.gz
9lug.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 9lug_validation.pdf.gz
9lug_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 9lug_full_validation.pdf.gz
9lug_full_validation.pdf.gz 9lug_validation.xml.gz
9lug_validation.xml.gz 9lug_validation.cif.gz
9lug_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/lu/9lug
https://data.pdbj.org/pub/pdb/validation_reports/lu/9lug ftp://data.pdbj.org/pub/pdb/validation_reports/lu/9lug
ftp://data.pdbj.org/pub/pdb/validation_reports/lu/9lug リンク
リンク 集合体
集合体
 要素
要素

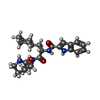
 タイプ: peptide-like, Peptide-like / クラス: 阻害剤 / 分子量: 452.546 Da / 分子数: 2 / 由来タイプ: 合成 / 式: C25H32N4O4 / タイプ: SUBJECT OF INVESTIGATION
タイプ: peptide-like, Peptide-like / クラス: 阻害剤 / 分子量: 452.546 Da / 分子数: 2 / 由来タイプ: 合成 / 式: C25H32N4O4 / タイプ: SUBJECT OF INVESTIGATION X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  SSRF
SSRF  / ビームライン: BL02U1 / 波長: 0.979183 Å
/ ビームライン: BL02U1 / 波長: 0.979183 Å 解析
解析 分子置換 / 解像度: 2.13→72.91 Å / SU ML: 0.23929363375 / 交差検証法: FREE R-VALUE / σ(F): 1.34009090092 / 位相誤差: 23.8898550139
分子置換 / 解像度: 2.13→72.91 Å / SU ML: 0.23929363375 / 交差検証法: FREE R-VALUE / σ(F): 1.34009090092 / 位相誤差: 23.8898550139  ムービー
ムービー コントローラー
コントローラー










 PDBj
PDBj
