[English] 日本語
 Yorodumi
Yorodumi- PDB-9h6i: Structure of the Arabidopsis thaliana 80S ribosome in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9h6i | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Arabidopsis thaliana 80S ribosome in complex with P- and E-site tRNAs, mRNA, and thermospermine | |||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||
 Keywords Keywords | RIBOSOME / plant / thermospermine / methylation / rRNA modification | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationroot morphogenesis / trichome morphogenesis / cytokinesis by cell plate formation / adaxial/abaxial pattern specification / leaf morphogenesis / response to high light intensity / plant-type cell wall / developmental process / chloroplast envelope / plasmodesma ...root morphogenesis / trichome morphogenesis / cytokinesis by cell plate formation / adaxial/abaxial pattern specification / leaf morphogenesis / response to high light intensity / plant-type cell wall / developmental process / chloroplast envelope / plasmodesma / plant-type vacuole / vacuole / response to UV-B / response to osmotic stress / nucleocytoplasmic transport / plastid / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / protein-RNA complex assembly / maturation of LSU-rRNA / response to salt stress / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / cytosolic ribosome / response to cold / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / chloroplast / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / translational initiation / small-subunit processome / modification-dependent protein catabolic process / protein tag activity / rRNA processing / peroxisome / ribosome biogenesis / ribosomal small subunit assembly / ribosomal small subunit biogenesis / response to oxidative stress / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / negative regulation of translation / defense response to bacterium / rRNA binding / structural constituent of ribosome / protein ubiquitination / ribosome / translation / ribonucleoprotein complex / cell division / mRNA binding / ubiquitin protein ligase binding / nucleolus / endoplasmic reticulum / mitochondrion / RNA binding / extracellular region / zinc ion binding / nucleoplasm / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||||||||||||||||||||
 Authors Authors | Faille, A. / Warren, A.J. | |||||||||||||||||||||||||||
| Funding support |  United Kingdom, European Union, 3items United Kingdom, European Union, 3items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2026 Journal: Science / Year: 2026Title: Structure of the Arabidopsis thaliana 80S ribosome in complex with P- and E-site tRNAs and mRNA Authors: Faille, A. / Warren, A.J. / Hellmann, E. / Ko, D. / Ruonala, R. / Helariutta, Y.E. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9h6i.cif.gz 9h6i.cif.gz | 5.4 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9h6i.ent.gz pdb9h6i.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  9h6i.json.gz 9h6i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/h6/9h6i https://data.pdbj.org/pub/pdb/validation_reports/h6/9h6i ftp://data.pdbj.org/pub/pdb/validation_reports/h6/9h6i ftp://data.pdbj.org/pub/pdb/validation_reports/h6/9h6i | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  51899MC  9h3gC  9hesC  9hmwC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: G / Beg label comp-ID: G / End auth comp-ID: A / End label comp-ID: A / Auth seq-ID: 1 - 76 / Label seq-ID: 1 - 76
NCS ensembles : (Details: Local NCS retraints between domains: 1 2) |
- Components
Components
-RNA chain , 2 types, 3 molecules 2W2B1
| #1: RNA chain | Mass: 24571.643 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #76: RNA chain | | Mass: 3629.032 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Ribosomal RNA ... , 4 types, 4 molecules 3AC3h1
| #2: RNA chain | Mass: 52827.414 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #3: RNA chain | Mass: 1096394.125 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #4: RNA chain | Mass: 38917.082 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #75: RNA chain | Mass: 582777.562 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
+Small ribosomal subunit protein ... , 31 types, 31 molecules BCMaAEJaVaKaACADDaPaXaBVBWNaABAAAFWaTaZaOaUaYaBBRaBLLaAaCaBaAI
+Large ribosomal subunit protein ... , 37 types, 37 molecules BMBOAUIaAXAPEaAWBDBSAMBIAHBTAVAJBQBHBKATBPBNBGFaHaBUBJAOAKBF...
-Protein , 2 types, 2 molecules ARGa
| #8: Protein | Mass: 9410.883 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #55: Protein | Mass: 14790.522 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Ribosomal protein ... , 2 types, 2 molecules ALBR
| #17: Protein | Mass: 25781.607 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #43: Protein | Mass: 28573.609 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein/peptide , 1 types, 1 molecules L3
| #79: Protein/peptide | Mass: 1975.426 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Non-polymers , 7 types, 11420 molecules 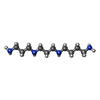












| #80: Chemical | ChemComp-TER / #81: Chemical | ChemComp-MG / #82: Chemical | ChemComp-K / #83: Chemical | ChemComp-SPD / | #84: Chemical | ChemComp-EPE / | #85: Chemical | ChemComp-ZN / #86: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Ribosome in complex with P- and E-site tRNAs, mRNA, polypeptide, and thermospermine Type: RIBOSOME / Entity ID: #1-#72, #79, #76-#78 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1800 nm / Nominal defocus min: 600 nm |
| Image recording | Electron dose: 40 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3D reconstruction | Resolution: 2.2 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 152921 / Symmetry type: POINT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 2.2→308.388 Å / Cor.coef. Fo:Fc: 0.787 / WRfactor Rwork: 0.354 / SU B: 7.38 / SU ML: 0.15 / Average fsc free: 0 / Average fsc overall: 0.7754 / Average fsc work: 0.7754 / ESU R: 0.188 Details: Hydrogens have been added in their riding positions
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: NONE | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 78.373 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj



































