[English] 日本語
 Yorodumi
Yorodumi- PDB-9h61: Auxin transporter-like protein 3 (LAX3) in the inward occluded st... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9h61 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Auxin transporter-like protein 3 (LAX3) in the inward occluded state in complex with auxin (Indole-3-acetic acid, IAA) | |||||||||
 Components Components | Auxin transporter-like protein 3 | |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Auxin transmembrane transport / AAAP family / APC superfamily | |||||||||
| Function / homology |  Function and homology information Function and homology informationroot cap development / lateral root formation / auxin influx transmembrane transporter activity / auxin polar transport / amino acid transmembrane transport / response to auxin / auxin-activated signaling pathway / amino acid transmembrane transporter activity / symporter activity / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Ung, K.L. / Andersen, C.G. / Stokes, D.L. / Pedersen, B.P. | |||||||||
| Funding support | European Union,  Denmark, 2items Denmark, 2items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2025 Journal: Nat Plants / Year: 2025Title: Structures and mechanism of the AUX/LAX transporters involved in auxin import. Authors: Kien Lam Ung / Lukas Schulz / Lorena Zuzic / Bjørn Lildal Amsinck / Sarah Koutnik-Abele / Ines Benhammouche / Camilla Gottlieb Andersen / Lynette Nel / Birgit Schiøtt / David L Stokes / ...Authors: Kien Lam Ung / Lukas Schulz / Lorena Zuzic / Bjørn Lildal Amsinck / Sarah Koutnik-Abele / Ines Benhammouche / Camilla Gottlieb Andersen / Lynette Nel / Birgit Schiøtt / David L Stokes / Ulrich Zeno Hammes / Bjørn Panyella Pedersen /     Abstract: Auxins are plant hormones that direct the growth and development of organisms on the basis of environmental cues. Indole-3-acetic acid (IAA) is the most abundant auxin in most plants. A variety of ...Auxins are plant hormones that direct the growth and development of organisms on the basis of environmental cues. Indole-3-acetic acid (IAA) is the most abundant auxin in most plants. A variety of membrane transport proteins work together to distribute auxins. These include the AUX/LAX protein family that mediate auxin import from the apoplast to the cytosol. Here we use structural and biophysical approaches combined with molecular dynamics to study transport by Arabidopsis thaliana LAX3, which is essential for plant root formation. Transport assays document high-affinity transport of IAA, as well as competitive behaviour of the synthetic phenoxyacetic acid auxin herbicide 2,4-dichlorophenoxyacetic acid and the auxin transport inhibitors 1-naphthoxyacetic acid and 2-naphthoxyacetic acid. Four cryo-EM structures were solved with resolutions of 2.9-3.4 Å: an inward open apo structure, two inward semi-occluded structures in complex with IAA and 2,4-dichlorophenoxyacetic acid, and a fully occluded structure in complex with 2-naphthoxyacetic acid. Structurally, LAX3 consists of a bundle and a scaffold domain. The ligand-binding site is sandwiched between these domains with two histidines occupying positions analogous to the sodium-binding sites in distantly related sodium:neurotransmitter transporters. This architecture suggests that these histidines couple transport to the proton motive force. Molecular dynamics simulations are used to explore substrate binding and release, including their dependence on specific protonation states. This study advances our understanding of auxin recognition and transport by AUX/LAX, providing insights into a fundamental aspect of plant physiology and development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9h61.cif.gz 9h61.cif.gz | 148.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9h61.ent.gz pdb9h61.ent.gz | 115.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9h61.json.gz 9h61.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/h6/9h61 https://data.pdbj.org/pub/pdb/validation_reports/h6/9h61 ftp://data.pdbj.org/pub/pdb/validation_reports/h6/9h61 ftp://data.pdbj.org/pub/pdb/validation_reports/h6/9h61 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  51894MC  9h62C  9h63C  9qqmC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 49442.750 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
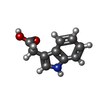
| #2: Chemical | ChemComp-IAC / |
| #3: Water | ChemComp-HOH / |
| Has ligand of interest | Y |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: LAX3 with IAA / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.049 MDa / Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  | ||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Details: 15 mA / Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K Details: Wait 4 seconds after sample loading, Blotting time 4 seconds with blotting force of -1 before plunging |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microscopy | Model: TFS KRIOS | |||||||||||||||||||||
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM | |||||||||||||||||||||
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 130000 X / Nominal defocus max: 2000 nm / Nominal defocus min: 600 nm / Cs: 2.7 mm / Alignment procedure: COMA FREE | |||||||||||||||||||||
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER | |||||||||||||||||||||
| Image recording |
| |||||||||||||||||||||
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV | |||||||||||||||||||||
| Image scans |
|
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: CTF amplitude correction was performed following motion correction using cryosparc-live Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 4432384 | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.21 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 74645 / Algorithm: FOURIER SPACE / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | B value: 111.9 / Space: REAL | ||||||||||||||||||||||||
| Atomic model building | Accession code: Q9CA25 / Source name: AlphaFold / Type: in silico model | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj