[English] 日本語
 Yorodumi
Yorodumi- PDB-9gef: Experimental localization of metal-binding sites reveals the role... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9gef | ||||||
|---|---|---|---|---|---|---|---|
| Title | Experimental localization of metal-binding sites reveals the role of metal ions in the delafloxacin-stabilized Streptococcus pneumoniae topoisomerase IV DNA cleavage complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN / metal ions / Type II topoisomerases / Fluoroquinolones / Long-wavelength X-ray crystallography | ||||||
| Function / homology | ACETATE ION / : / delafloxacin / DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Biological species |   | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.62 Å MOLECULAR REPLACEMENT / Resolution: 2.62 Å | ||||||
 Authors Authors | Wang, B. / Najmudin, S. / Pan, X.-S. / Mykhaylyk, V. / Orr, C. / Wagner, A. / Govada, L. / Chayen, N.E. / Fisher, L.M. / Sanderson, M.R. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2024 Journal: Proc.Natl.Acad.Sci.USA / Year: 2024Title: Experimental localization of metal-binding sites reveals the role of metal ions in type II DNA topoisomerases. Authors: Wang, B. / Najmudin, S. / Pan, X.S. / Mykhaylyk, V. / Orr, C. / Wagner, A. / Govada, L. / Chayen, N.E. / Fisher, L.M. / Sanderson, M.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9gef.cif.gz 9gef.cif.gz | 627.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9gef.ent.gz pdb9gef.ent.gz | 512.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9gef.json.gz 9gef.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ge/9gef https://data.pdbj.org/pub/pdb/validation_reports/ge/9gef ftp://data.pdbj.org/pub/pdb/validation_reports/ge/9gef ftp://data.pdbj.org/pub/pdb/validation_reports/ge/9gef | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 81934.758 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: THE FUSED TOPOISOMERASE IV CLEAVAGE COMPLEX COMPRISES THE C-TERMINAL DOMAIN OF THE PARE30 DOMAIN (RESIDUES 415-647), A HIS INSERT AT POSITION 648 AND THE N-TERMINAL DOMAIN OF PARC55 (RESIDUES 1001-1486), Source: (gene. exp.)   |
|---|
-DNA chain , 4 types, 4 molecules EFGH
| #2: DNA chain | Mass: 2168.445 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: PLAMID PBR322 / Source: (synth.)  |
|---|---|
| #3: DNA chain | Mass: 3333.198 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: PLASMID PBR322 / Source: (synth.)  |
| #4: DNA chain | Mass: 2121.436 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: PLAMID PBR322 / Source: (synth.)  |
| #5: DNA chain | Mass: 3317.199 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: PLAMID PBR322 / Source: (synth.)  |
-Non-polymers , 7 types, 129 molecules 




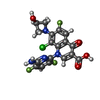







| #6: Chemical | ChemComp-MPD / ( #7: Chemical | ChemComp-MG / #8: Chemical | ChemComp-K / #9: Chemical | #10: Chemical | ChemComp-ACT / | #11: Chemical | #12: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | N |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.37 Å3/Da / Density % sol: 71.88 % / Description: rod-shaped |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: 50 mM sodium cacodylate, 62.5 mM KCl, 7.5 mM MgCl 2 , 2.5% Tacsimate TM Hampton Research, 5.5-7% isopropanol, pH 6.5. The protein crystals were cryoprotected with 50 mM sodium cacodylate pH ...Details: 50 mM sodium cacodylate, 62.5 mM KCl, 7.5 mM MgCl 2 , 2.5% Tacsimate TM Hampton Research, 5.5-7% isopropanol, pH 6.5. The protein crystals were cryoprotected with 50 mM sodium cacodylate pH 6.5, 62.5 mM KCl, 7.5 mM MgCl 2 , 2.5% Tacsimate TM Hampton Research, 1 mM beta-mercaptoethanol and 30% v/v MPD before being flash-cooled in liquid nitrogen and collected in elliptical polyimide sample mounts. PH range: 6.5 - 8.0 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I23 / Wavelength: 2.75,3.14,3.54,4.35,4.50,5.16 / Beamline: I23 / Wavelength: 2.75,3.14,3.54,4.35,4.50,5.16 | |||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 12M / Detector: PIXEL / Date: Sep 23, 2023 | |||||||||||||||||||||
| Radiation | Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||
| Radiation wavelength |
| |||||||||||||||||||||
| Reflection | Resolution: 2.37→210.82 Å / Num. obs: 124385 / % possible obs: 100 % / Redundancy: 59.4 % / CC1/2: 0.999 / Rmerge(I) obs: 0.215 / Rpim(I) all: 0.027 / Net I/σ(I): 11.6 | |||||||||||||||||||||
| Reflection shell | Resolution: 2.37→2.41 Å / Redundancy: 45 % / Rmerge(I) obs: 5.369 / Num. unique obs: 6112 / CC1/2: 0.204 / Rpim(I) all: 0.796 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.62→79.39 Å / Cor.coef. Fo:Fc: 0.942 / Cor.coef. Fo:Fc free: 0.926 / SU B: 26.622 / SU ML: 0.23 / Cross valid method: THROUGHOUT / ESU R: 0.524 / ESU R Free: 0.276 / Stereochemistry target values: MAXIMUM LIKELIHOOD MOLECULAR REPLACEMENT / Resolution: 2.62→79.39 Å / Cor.coef. Fo:Fc: 0.942 / Cor.coef. Fo:Fc free: 0.926 / SU B: 26.622 / SU ML: 0.23 / Cross valid method: THROUGHOUT / ESU R: 0.524 / ESU R Free: 0.276 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: U VALUES : WITH TLS ADDED HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT U VALUES : RESIDUAL ONLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.1 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 48.179 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.62→79.39 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.622→2.69 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj












































