[English] 日本語
 Yorodumi
Yorodumi- PDB-9di6: Crystal structure of Plasmodium falciparum dihydroorotate dehydro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9di6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM679 (ethyl 1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxylate) | ||||||
 Components Components | Dihydroorotate dehydrogenase (quinone), mitochondrial | ||||||
 Keywords Keywords | OXIDOREDUCTASE/INHIBITOR / inhibitor / Alpha-Beta Barrel / OXIDOREDUCTASE / Flavoprotein / OXIDOREDUCTASE-INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationPyrimidine biosynthesis / pyrimidine ribonucleotide biosynthetic process / dihydroorotate dehydrogenase (quinone) / dihydroorotate dehydrogenase (quinone) activity / dihydroorotate dehydrogenase activity / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process / mitochondrial inner membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.41 Å MOLECULAR REPLACEMENT / Resolution: 2.41 Å | ||||||
 Authors Authors | Deng, X. / Tomchick, D. / Phillips, M. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2025 Journal: J.Med.Chem. / Year: 2025Title: Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention. Authors: Nie, Z. / Bonnert, R. / Tsien, J. / Deng, X. / Higgs, C. / El Mazouni, F. / Zhang, X. / Li, R. / Ho, N. / Feher, V. / Paulsen, J. / Shackleford, D.M. / Katneni, K. / Chen, G. / Ng, A.C.F. / ...Authors: Nie, Z. / Bonnert, R. / Tsien, J. / Deng, X. / Higgs, C. / El Mazouni, F. / Zhang, X. / Li, R. / Ho, N. / Feher, V. / Paulsen, J. / Shackleford, D.M. / Katneni, K. / Chen, G. / Ng, A.C.F. / McInerney, M. / Wang, W. / Saunders, J. / Collins, D. / Yan, D. / Li, P. / Campbell, M. / Patil, R. / Ghoshal, A. / Mondal, P. / Kundu, A. / Chittimalla, R. / Mahadeva, M. / Kokkonda, S. / White, J. / Das, R. / Mukherjee, P. / Angulo-Barturen, I. / Jimenez-Diaz, M.B. / Malmstrom, R. / Lawrenz, M. / Rodriguez-Granillo, A. / Rathod, P.K. / Tomchick, D.R. / Palmer, M.J. / Laleu, B. / Qin, T. / Charman, S.A. / Phillips, M.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9di6.cif.gz 9di6.cif.gz | 282.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9di6.ent.gz pdb9di6.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  9di6.json.gz 9di6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/di/9di6 https://data.pdbj.org/pub/pdb/validation_reports/di/9di6 ftp://data.pdbj.org/pub/pdb/validation_reports/di/9di6 ftp://data.pdbj.org/pub/pdb/validation_reports/di/9di6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  9dikC  9dizC  9dkoC  9dkqC  9dkyC  9dlkC  9dlyC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 45385.914 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q08210, dihydroorotate dehydrogenase (quinone) |
|---|
-Non-polymers , 6 types, 41 molecules 
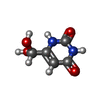







| #2: Chemical | ChemComp-A1A51 / Mass: 327.302 Da / Num. of mol.: 1 / Source method: obtained synthetically / Formula: C15H16F3N3O2 / Feature type: SUBJECT OF INVESTIGATION |
|---|---|
| #3: Chemical | ChemComp-FMN / |
| #4: Chemical | ChemComp-OG6 / |
| #5: Chemical | ChemComp-CIT / |
| #6: Chemical | ChemComp-LDA / |
| #7: Water | ChemComp-HOH / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.84 Å3/Da / Density % sol: 56.7 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 4.1 Details: 12% Ethylene Glycol, 0.13M Citric Acid pH 4.1, 9% PEG6000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.97924 Å / Beamline: 19-ID / Wavelength: 0.97924 Å |
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Mar 14, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97924 Å / Relative weight: 1 |
| Reflection | Resolution: 2.4→50 Å / Num. obs: 21869 / % possible obs: 99.9 % / Redundancy: 17.8 % / Biso Wilson estimate: 37.52 Å2 / CC1/2: 0.964 / Net I/σ(I): 35.26 |
| Reflection shell | Resolution: 2.4→2.44 Å / Redundancy: 16.4 % / Mean I/σ(I) obs: 1.25 / Num. unique obs: 1060 / CC1/2: 0.551 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.41→45.79 Å / SU ML: 0.2486 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 24.8955 MOLECULAR REPLACEMENT / Resolution: 2.41→45.79 Å / SU ML: 0.2486 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 24.8955 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 51.14 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.41→45.79 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Refine-ID: X-RAY DIFFRACTION / Auth asym-ID: A / Label asym-ID: A
|
 Movie
Movie Controller
Controller


 PDBj
PDBj



