+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9cus | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BmrCD in the outward-facing conformation bound to Hoechsts | |||||||||||||||||||||
 Components Components | (Probable multidrug resistance ABC transporter ATP-binding/permease protein ...) x 2 | |||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / ABC transporter / Exporter / transport | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationATPase-coupled lipid transmembrane transporter activity / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / transmembrane transport / response to antibiotic / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||||||||
 Authors Authors | Tang, Q. / Mchaourab, H.S. | |||||||||||||||||||||
| Funding support |  United States, 1items United States, 1items
| |||||||||||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2025 Journal: bioRxiv / Year: 2025Title: Lipid-mediated mechanism of drug extrusion by a heterodimeric ABC exporter. Authors: Qingyu Tang / Matt Sinclair / Paola Bisignano / Yunsen Zhang / Emad Tajkhorshid / Hassane S Mchaourab /  Abstract: Multidrug transport by ATP binding cassette (ABC) exporters entails a mechanism to modulate drug affinity across the transport cycle. Here, we combine cryo-EM and molecular dynamics (MD) simulations ...Multidrug transport by ATP binding cassette (ABC) exporters entails a mechanism to modulate drug affinity across the transport cycle. Here, we combine cryo-EM and molecular dynamics (MD) simulations to illuminate how lipid competition modulates substrate affinity to drive its translocation by ABC exporters. We determined cryo-EM structures of the ABC transporter BmrCD in drug-loaded inward-facing (IF) and outward-facing (OF) conformations in lipid nanodiscs to reveal the structural basis of alternating access, details of drug-transporter interactions, and the scale of drug movement between the two conformations. Remarkably, the structures uncovered lipid molecules bound in or near the transporter vestibule along with the drugs. MD trajectories from the IF structure show that these lipids stimulate drug disorder and translocation towards the vestibule apex. Similarly, bound lipids enter the OF vestibule and weaken drug-transporter interactions facilitating drug release. Our results complete a near-atomic model of BmrCD's conformational cycle and advance a general mechanism of lipid-driven drug transport by ABC exporters. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9cus.cif.gz 9cus.cif.gz | 525.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9cus.ent.gz pdb9cus.ent.gz | 440.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9cus.json.gz 9cus.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  9cus_validation.pdf.gz 9cus_validation.pdf.gz | 1.5 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  9cus_full_validation.pdf.gz 9cus_full_validation.pdf.gz | 1.5 MB | Display | |
| Data in XML |  9cus_validation.xml.gz 9cus_validation.xml.gz | 43.3 KB | Display | |
| Data in CIF |  9cus_validation.cif.gz 9cus_validation.cif.gz | 63.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cu/9cus https://data.pdbj.org/pub/pdb/validation_reports/cu/9cus ftp://data.pdbj.org/pub/pdb/validation_reports/cu/9cus ftp://data.pdbj.org/pub/pdb/validation_reports/cu/9cus | HTTPS FTP |
-Related structure data
| Related structure data |  45940MC  9cupC  9curC  46002  46003 M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Probable multidrug resistance ABC transporter ATP-binding/permease protein ... , 2 types, 2 molecules CD
| #1: Protein | Mass: 67589.922 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: yheI, BSU09710 Production host:  References: UniProt: O07550, Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| #2: Protein | Mass: 77467.078 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: yheH, BSU09720 Production host:  References: UniProt: O07549, Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
-Non-polymers , 5 types, 26 molecules 



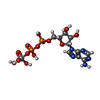




| #3: Chemical | | #4: Chemical | ChemComp-ATP / | #5: Chemical | #6: Chemical | ChemComp-POV / ( #7: Chemical | ChemComp-AOV / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Heterodimeric BmrCD with ligands ATP and lipids / Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 3 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2200 nm / Nominal defocus min: 400 nm / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 15 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.21.1_5286: / Classification: refinement |
|---|---|
| EM software | Name: PHENIX / Category: model refinement |
| CTF correction | Type: NONE |
| 3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 27802 / Symmetry type: POINT |
 Movie
Movie Controller
Controller









 PDBj
PDBj






