| Entry | Database: PDB / ID: 9cuo
|
|---|
| Title | Crystal structure of CRBN with compound 3 |
|---|
 Components Components | Protein cereblon |
|---|
 Keywords Keywords | LIGASE / CRBN / E3 / protein degradation |
|---|
| Function / homology |  Function and homology information Function and homology information
negative regulation of monoatomic ion transmembrane transport / Cul4A-RING E3 ubiquitin ligase complex / locomotory exploration behavior / positive regulation of Wnt signaling pathway / negative regulation of protein-containing complex assembly / positive regulation of protein-containing complex assembly / Potential therapeutics for SARS / proteasome-mediated ubiquitin-dependent protein catabolic process / transmembrane transporter binding / protein ubiquitination ...negative regulation of monoatomic ion transmembrane transport / Cul4A-RING E3 ubiquitin ligase complex / locomotory exploration behavior / positive regulation of Wnt signaling pathway / negative regulation of protein-containing complex assembly / positive regulation of protein-containing complex assembly / Potential therapeutics for SARS / proteasome-mediated ubiquitin-dependent protein catabolic process / transmembrane transporter binding / protein ubiquitination / perinuclear region of cytoplasm / metal ion binding / nucleus / membrane / cytosol / cytoplasmSimilarity search - Function Yippee/Mis18/Cereblon / Yippee zinc-binding/DNA-binding /Mis18, centromere assembly / CULT domain / CULT domain profile. / Lon N-terminal domain profile. / Lon protease, N-terminal domain / Lon protease, N-terminal domain superfamily / ATP-dependent protease La (LON) substrate-binding domain / Found in ATP-dependent protease La (LON) / PUA-like superfamilySimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å |
|---|
 Authors Authors | Zheng, X. / Ji, N. / Campbell, V. / Slavin, A. / Zhu, X. / Chen, D. / Rong, H. / Enerson, B. / Mayo, M. / Sharma, K. ...Zheng, X. / Ji, N. / Campbell, V. / Slavin, A. / Zhu, X. / Chen, D. / Rong, H. / Enerson, B. / Mayo, M. / Sharma, K. / Browne, C.M. / Klaus, C.R. / Li, H. / Massa, G. / McDonald, A.A. / Shi, Y. / Sintchak, M. / Skouras, S. / Walther, D.M. / Yuan, K. / Zhang, Y. / Kelleher, J. / Guang, L. / Luo, X. / Mainolfi, N. / Weiss, M.M. |
|---|
| Funding support |  United States, 1items United States, 1items | Organization | Grant number | Country |
|---|
| Other private | |  United States United States |
|
|---|
 Citation Citation |  Journal: J.Med.Chem. / Year: 2024 Journal: J.Med.Chem. / Year: 2024
Title: Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
Authors: Zheng, X. / Ji, N. / Campbell, V. / Slavin, A. / Zhu, X. / Chen, D. / Rong, H. / Enerson, B. / Mayo, M. / Sharma, K. / Browne, C.M. / Klaus, C.R. / Li, H. / Massa, G. / McDonald, A.A. / Shi, ...Authors: Zheng, X. / Ji, N. / Campbell, V. / Slavin, A. / Zhu, X. / Chen, D. / Rong, H. / Enerson, B. / Mayo, M. / Sharma, K. / Browne, C.M. / Klaus, C.R. / Li, H. / Massa, G. / McDonald, A.A. / Shi, Y. / Sintchak, M. / Skouras, S. / Walther, D.M. / Yuan, K. / Zhang, Y. / Kelleher, J. / Liu, G. / Luo, X. / Mainolfi, N. / Weiss, M.M. |
|---|
| History | | Deposition | Jul 26, 2024 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Aug 28, 2024 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Nov 6, 2024 | Group: Database references / Structure summary / Category: citation / citation_author / pdbx_entry_details
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation_author.identifier_ORCID / _pdbx_entry_details.has_protein_modification |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å
MOLECULAR REPLACEMENT / Resolution: 1.6 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: J.Med.Chem. / Year: 2024
Journal: J.Med.Chem. / Year: 2024 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 9cuo.cif.gz
9cuo.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb9cuo.ent.gz
pdb9cuo.ent.gz PDB format
PDB format 9cuo.json.gz
9cuo.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 9cuo_validation.pdf.gz
9cuo_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 9cuo_full_validation.pdf.gz
9cuo_full_validation.pdf.gz 9cuo_validation.xml.gz
9cuo_validation.xml.gz 9cuo_validation.cif.gz
9cuo_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/cu/9cuo
https://data.pdbj.org/pub/pdb/validation_reports/cu/9cuo ftp://data.pdbj.org/pub/pdb/validation_reports/cu/9cuo
ftp://data.pdbj.org/pub/pdb/validation_reports/cu/9cuo F&H Search
F&H Search Links
Links Assembly
Assembly
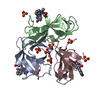

 Components
Components Homo sapiens (human) / Gene: CRBN, AD-006 / Production host:
Homo sapiens (human) / Gene: CRBN, AD-006 / Production host: 







 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  CLSI
CLSI  / Beamline: 08ID-1 / Wavelength: 1.0332 Å
/ Beamline: 08ID-1 / Wavelength: 1.0332 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 1.6→42.72 Å / Cor.coef. Fo:Fc: 0.973 / Cor.coef. Fo:Fc free: 0.957 / SU B: 3.072 / SU ML: 0.05 / Cross valid method: THROUGHOUT / ESU R: 0.089 / ESU R Free: 0.081 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MOLECULAR REPLACEMENT / Resolution: 1.6→42.72 Å / Cor.coef. Fo:Fc: 0.973 / Cor.coef. Fo:Fc free: 0.957 / SU B: 3.072 / SU ML: 0.05 / Cross valid method: THROUGHOUT / ESU R: 0.089 / ESU R Free: 0.081 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller



 PDBj
PDBj


