[English] 日本語
 Yorodumi
Yorodumi- PDB-9cq0: Event-based electron counting microED structure of thiostrepton f... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9cq0 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Event-based electron counting microED structure of thiostrepton from a single crystal | ||||||||||||
 Components Components | Thiostrepton | ||||||||||||
 Keywords Keywords | ANTIBIOTIC / Cyclic / macrocycle | ||||||||||||
| Function / homology | THIOSTREPTON Function and homology information Function and homology information | ||||||||||||
| Biological species |  Streptomyces azureus (bacteria) Streptomyces azureus (bacteria) | ||||||||||||
| Method | ELECTRON CRYSTALLOGRAPHY / electron crystallography / Resolution: 1.5 Å | ||||||||||||
 Authors Authors | Vlahakis, N.W. / Qu, S. / Richards, L.S. / deMoraes, L.S. / Nelson, H.M. / Rodriguez, J.A. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: Acta Crystallogr C Struct Chem / Year: 2025 Journal: Acta Crystallogr C Struct Chem / Year: 2025Title: Fast event-based electron counting for small-molecule structure determination by MicroED. Authors: Niko Vlahakis / Songrong Qu / Logan S Richards / Lygia Silva de Moraes / Duilio Cascio / Hosea M Nelson / Jose A Rodriguez /  Abstract: Electron counting helped realize the resolution revolution in single-particle cryoEM and is now accelerating the determination of MicroED structures. Its advantages are best demonstrated by new ...Electron counting helped realize the resolution revolution in single-particle cryoEM and is now accelerating the determination of MicroED structures. Its advantages are best demonstrated by new direct electron detectors capable of fast (kilohertz) event-based electron counting (EBEC). This strategy minimizes the inaccuracies introduced by coincidence loss (CL) and promises rapid determination of accurate structures. We used the Direct Electron Apollo camera to leverage EBEC technology for MicroED data collection. Given its ability to count single electrons, the Apollo collects high-quality MicroED data from organic small-molecule crystals illuminated with incident electron beam flux densities as low as 0.01-0.045 e/Å/s. Under even the lowest flux density (0.01 e/Å/s) condition, fast EBEC data produced ab initio structures of a salen ligand (268 Da) and biotin (244 Da). Each structure was determined from a 100° wedge of data collected from a single crystal in as few as 50 s, with a delivered fluence of only ∼0.5 e/Å. Fast EBEC data collected with a fluence of 2.25 or 3.33 e/Å also facilitated a 1.5 Å structure of thiostrepton (1665 Da). While refinement of these structures appeared unaffected by CL, a CL adjustment applied to EBEC data further improved the distribution of intensities measured from the salen ligand and biotin crystals. However, CL adjustment only marginally improved the refinement of their corresponding structures, signaling the already high counting accuracy of detectors with counting rates in the kilohertz range. Overall, by delivering low-dose structure-worthy data, fast EBEC collection strategies open new possibilities for high-throughput MicroED. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9cq0.cif.gz 9cq0.cif.gz | 18.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9cq0.ent.gz pdb9cq0.ent.gz | 10.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9cq0.json.gz 9cq0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cq/9cq0 https://data.pdbj.org/pub/pdb/validation_reports/cq/9cq0 ftp://data.pdbj.org/pub/pdb/validation_reports/cq/9cq0 ftp://data.pdbj.org/pub/pdb/validation_reports/cq/9cq0 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
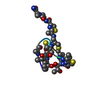
| #1: Protein/peptide | 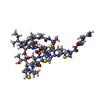  Type: Thiopeptide / Class: Antibiotic / Mass: 1805.985 Da / Num. of mol.: 1 / Source method: isolated from a natural source Type: Thiopeptide / Class: Antibiotic / Mass: 1805.985 Da / Num. of mol.: 1 / Source method: isolated from a natural sourceDetails: Thiostrepton is a hetrocyclic thiopeptide belonging to the thiocillin family, consisting of four thiazole, one thiozoline and one piperideine rings. A modified quinoline linked to main-chain ...Details: Thiostrepton is a hetrocyclic thiopeptide belonging to the thiocillin family, consisting of four thiazole, one thiozoline and one piperideine rings. A modified quinoline linked to main-chain residue 1 and side-chain of residue 12. Post translational maturation of thiazole and oxazole containing antibiotics involves the enzymic condensation of a Cys or Ser with the alpha-carbonyl of the preceding amino acid to form a thioether or ether bond, then dehydration to form a double bond with the alpha-amino nitrogen. Thiazoline or oxazoline ring are dehydrogenated to form thiazole or oxazole rings. the pyridinyl involves the cross-linking of a Ser and a Cys-Ser pair usually separated by 7 or 8 residues along the peptide chain. The Ser residues are dehydrated to didehydroalanines, then bonded between their beta carbons. The alpha carbonyl of the Cys condenses with alpha carbon of the first Ser to form a pyridinyl ring. The ring may be mutiply dehydrogenated to form a pyridine ring with loss of the amino nitrogen of the first Ser. The amidation of Ser-17 probably does not occur by the same mechanism, oxidative cleavage of glycine, as in eukaryotes. Source: (natural)  Streptomyces azureus (bacteria) / References: THIOSTREPTON Streptomyces azureus (bacteria) / References: THIOSTREPTON |
|---|---|
| #2: Water | ChemComp-HOH / |
| Compound details | THIOSTREPTON IS A MEMBER OF A SULPHUR-RICH HETEROCYCLIC PEPTIDES CLASS. ALL SHARE A MACROCYLIC ...THIOSTREPT |
| Has ligand of interest | N |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON CRYSTALLOGRAPHY |
|---|---|
| EM experiment | Aggregation state: 3D ARRAY / 3D reconstruction method: electron crystallography |
- Sample preparation
Sample preparation
| Component | Name: Microcrystal of thiostrepton / Type: COMPLEX / Entity ID: #1 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 1.81 kDa/nm / Experimental value: NO |
| Source (natural) | Organism:  Streptomyces azureus (bacteria) Streptomyces azureus (bacteria) |
| EM crystal formation | Atmosphere: Standard temperature and pressure Details: Thiostrepton (30 mg) was dissolved in 1.95 mL 24:1 Chloroform:Isoamyl Alcohol. 390 uL of Ethanol and 195 uL of 100% Glycerol were each mixed into the solution, and tetragonal crystals of ...Details: Thiostrepton (30 mg) was dissolved in 1.95 mL 24:1 Chloroform:Isoamyl Alcohol. 390 uL of Ethanol and 195 uL of 100% Glycerol were each mixed into the solution, and tetragonal crystals of various sizes (including microcrystals) formed after approximately 2 days of slow evaporation of the solvent under ambient conditions. Temperature: 293 K / Time: 2 DAY |
| Buffer solution | pH: 7 / Details: 24:1 chloroform/isoamyl alcohol |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: NO |
-Data collection
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TALOS ARCTICA |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: DIFFRACTION / Nominal defocus max: 0 nm / Nominal defocus min: 0 nm / C2 aperture diameter: 70 µm |
| Specimen holder | Cryogen: NITROGEN / Temperature (max): 100 K / Temperature (min): 100 K |
| Image recording | Average exposure time: 3.33 sec. / Electron dose: 0.0333 e/Å2 / Film or detector model: DIRECT ELECTRON APOLLO (4k x 4k) / Num. of diffraction images: 100 / Num. of grids imaged: 1 |
| Image scans | Width: 4096 / Height: 4096 |
| EM diffraction shell | Resolution: 1.5→1.6 Å / Fourier space coverage: 98.6 % / Multiplicity: 7.4 / Num. of structure factors: 289 / Phase residual: 38 ° |
| EM diffraction stats | Fourier space coverage: 99.3 % / High resolution: 1.5 Å / Num. of intensities measured: 12605 / Num. of structure factors: 1738 Phase error rejection criteria: Phases were determined by molecular replacement and cross-validated by free R-value during refinement Rmerge: 20.4 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Image processing | Details: Images were gain corrected and binned at 4k x 4k pixels. A pedestal of 1 pixel value was applied to each pixel during conversion to SMV file format. | ||||||||||||||||||||||||
| EM 3D crystal entity | ∠α: 90 ° / ∠β: 90 ° / ∠γ: 90 ° / A: 26.47 Å / B: 26.47 Å / C: 27.29 Å / Space group name: P4(3)2(1)2 / Space group num: 96 | ||||||||||||||||||||||||
| CTF correction | Type: NONE | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 1.5 Å / Resolution method: DIFFRACTION PATTERN/LAYERLINES / Symmetry type: 3D CRYSTAL | ||||||||||||||||||||||||
| Atomic model building | B value: 13.09 | ||||||||||||||||||||||||
| Atomic model building | PDB-ID: 1E9W Accession code: 1E9W / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||
| Refinement | Resolution: 1.5→18.72 Å / SU ML: 0.07 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 16.39 / Stereochemistry target values: ML
| ||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.5→18.72 Å
|
 Movie
Movie Controller
Controller


 PDBj
PDBj