[English] 日本語
 Yorodumi
Yorodumi- PDB-9cjf: CryoEM structure of alkaline-inactivated nitrogenase MoFe-protein... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9cjf | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of alkaline-inactivated nitrogenase MoFe-protein in complex with NafT | ||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||
 Keywords Keywords | METAL BINDING PROTEIN / Oxidoreductase | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | ||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.33 Å | ||||||||||||||||||||||||
 Authors Authors | Warmack, R.A. / Rees, D.C. | ||||||||||||||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural evolution of nitrogenase states under alkaline turnover. Authors: Rebeccah A Warmack / Douglas C Rees /  Abstract: Biological nitrogen fixation, performed by the enzyme nitrogenase, supplies nearly 50% of the bioavailable nitrogen pool on Earth, yet the structural nature of the enzyme intermediates involved in ...Biological nitrogen fixation, performed by the enzyme nitrogenase, supplies nearly 50% of the bioavailable nitrogen pool on Earth, yet the structural nature of the enzyme intermediates involved in this cycle remains ambiguous. Here we present four high resolution cryoEM structures of the nitrogenase MoFe-protein, sampled along a time course of alkaline reaction mixtures under an acetylene atmosphere. This series of structures reveals a sequence of salient changes including perturbations to the inorganic framework of the FeMo-cofactor; depletion of the homocitrate moiety; diminished density around the S2B belt sulfur of the FeMo-cofactor; rearrangements of cluster-adjacent side chains; and the asymmetric displacement of the FeMo-cofactor. We further demonstrate that the nitrogenase associated factor T protein can recognize and bind an alkaline inactivated MoFe-protein in vitro. These time-resolved structures provide experimental support for the displacement of S2B and distortions of the FeMo-cofactor at the E-E intermediates of the substrate reduction mechanism, prior to nitrogen binding, highlighting cluster rearrangements potentially relevant to nitrogen fixation by biological and synthetic clusters. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9cjf.cif.gz 9cjf.cif.gz | 402.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9cjf.ent.gz pdb9cjf.ent.gz | 321.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9cjf.json.gz 9cjf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cj/9cjf https://data.pdbj.org/pub/pdb/validation_reports/cj/9cjf ftp://data.pdbj.org/pub/pdb/validation_reports/cj/9cjf ftp://data.pdbj.org/pub/pdb/validation_reports/cj/9cjf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  45630MC  9cjbC  9cjcC  9cjdC  9cjeC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Nitrogenase molybdenum-iron protein ... , 2 types, 4 molecules ACBD
| #1: Protein | Mass: 55363.043 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase Azotobacter vinelandii (bacteria) / References: UniProt: P07328, nitrogenase#2: Protein | Mass: 59535.879 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase Azotobacter vinelandii (bacteria) / References: UniProt: P07329, nitrogenase |
|---|
-Protein , 1 types, 1 molecules E
| #3: Protein | Mass: 15154.773 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azotobacter vinelandii (bacteria) / Gene: Avin_01560 / Production host: Azotobacter vinelandii (bacteria) / Gene: Avin_01560 / Production host:  |
|---|
-Non-polymers , 5 types, 720 molecules 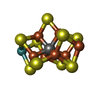







| #4: Chemical | | #5: Chemical | #6: Chemical | #7: Chemical | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Heterotetrameric molybdenum-iron protein in complex with one NafT molecule Type: COMPLEX / Entity ID: #1-#3 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 0.247 MDa / Experimental value: NO |
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
| Source (recombinant) | Organism: unidentified (others) |
| Buffer solution | pH: 7.8 |
| Specimen | Conc.: 6 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 3000 nm / Nominal defocus min: 600 nm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement |
|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| 3D reconstruction | Resolution: 2.33 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 78918 / Symmetry type: POINT |
 Movie
Movie Controller
Controller






 PDBj
PDBj




