+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9c9m | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 intasome core bound with DTG | |||||||||||||||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | VIRAL PROTEIN / HIV-1 / integrase / nucleoprotein complexes / INSTI / intasome / CryoEM | |||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationHIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding ...HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / host cell / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.01 Å | |||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Li, M. / Craigie, R. | |||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 1items United States, 1items
| |||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2024 Journal: Viruses / Year: 2024Title: HIV-1 Intasomes Assembled with Excess Integrase C-Terminal Domain Protein Facilitate Structural Studies by Cryo-EM and Reveal the Role of the Integrase C-Terminal Tail in HIV-1 Integration. Authors: Min Li / Zhen Li / Xuemin Chen / Yanxiang Cui / Alan N Engelman / Robert Craigie /   Abstract: Retroviral integration is mediated by intasome nucleoprotein complexes wherein a pair of viral DNA ends are bridged together by a multimer of integrase (IN). Atomic-resolution structures of HIV-1 ...Retroviral integration is mediated by intasome nucleoprotein complexes wherein a pair of viral DNA ends are bridged together by a multimer of integrase (IN). Atomic-resolution structures of HIV-1 intasomes provide detailed insights into the mechanism of integration and inhibition by clinical IN inhibitors. However, previously described HIV-1 intasomes are highly heterogeneous and have the tendency to form stacks, which is a limiting factor in determining high-resolution cryo-EM maps. We have assembled HIV-1 intasomes in the presence of excess IN C-terminal domain protein, which was readily incorporated into the intasomes. The purified intasomes were largely homogeneous and exhibited minimal stacking tendencies. The cryo-EM map resolution was further improved to 2.01 Å, which will greatly facilitate structural studies of IN inhibitor action and drug resistance mechanisms. The C-terminal 18 residues of HIV-1 IN, which are critical for virus replication and integration in vitro, have not been well resolved in previous intasome structures, and its function remains unclear. We show that the C-terminal tail participates in intasome assembly, resides within the intasome core, and forms a small alpha helix (residues 271-276). Mutations that disrupt alpha helix integrity impede IN activity in vitro and disrupt HIV-1 infection at the step of viral DNA integration. | |||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9c9m.cif.gz 9c9m.cif.gz | 313.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9c9m.ent.gz pdb9c9m.ent.gz | 238.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9c9m.json.gz 9c9m.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c9/9c9m https://data.pdbj.org/pub/pdb/validation_reports/c9/9c9m ftp://data.pdbj.org/pub/pdb/validation_reports/c9/9c9m ftp://data.pdbj.org/pub/pdb/validation_reports/c9/9c9m | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  45364MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 8 molecules ABIKLCDM
| #1: Protein | Mass: 39864.641 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Gene: gag-pol / Production host: Human immunodeficiency virus 1 / Gene: gag-pol / Production host:  References: UniProt: P12497, Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases, Hydrolases; Acting on ester bonds #2: Protein | Mass: 12101.872 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Gene: gag-pol / Production host: Human immunodeficiency virus 1 / Gene: gag-pol / Production host:  References: UniProt: P12497, Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases, Hydrolases; Acting on ester bonds |
|---|
-DNA chain , 2 types, 4 molecules ENFO
| #3: DNA chain | Mass: 8188.271 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1#4: DNA chain | Mass: 7773.023 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
|---|
-Non-polymers , 4 types, 89 molecules 

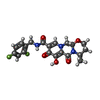




| #5: Chemical | ChemComp-MG / #6: Chemical | #7: Chemical | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: HIV-1 intasomes Core / Type: COMPLEX Details: HIV-1 intasomes assembled with full length integrase and integrase c-terminal domain Entity ID: #1-#4 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 6.2 |
| Specimen | Conc.: 0.4 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 105000 X / Nominal defocus max: 2600 nm / Nominal defocus min: 600 nm / Cs: 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 54 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| EM software |
| |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | |||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 940736 | |||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.01 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 428779 / Symmetry type: POINT | |||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | |||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 5U1C Accession code: 5U1C / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller



 PDBj
PDBj











































