[English] 日本語
 Yorodumi
Yorodumi- PDB-8yaz: XFEL crystal structure of the oxidized form of F87A/F393H P450BM3... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8yaz | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | XFEL crystal structure of the oxidized form of F87A/F393H P450BM3 with N-enanthyl-L-prolyl-L-phenylalanine in complex with styrene | |||||||||||||||
 Components Components | Bifunctional cytochrome P450/NADPH--P450 reductase | |||||||||||||||
 Keywords Keywords | OXIDOREDUCTASE / Cytochrome P450 | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationaromatase activity / NADPH-hemoprotein reductase / NADPH-hemoprotein reductase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen / unspecific monooxygenase / FMN binding / flavin adenine dinucleotide binding / iron ion binding / heme binding / identical protein binding / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) | |||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  FREE ELECTRON LASER / FREE ELECTRON LASER /  MOLECULAR REPLACEMENT / Resolution: 1.85 Å MOLECULAR REPLACEMENT / Resolution: 1.85 Å | |||||||||||||||
 Authors Authors | Nagao, S. / Kuwano, W. / Tosha, T. / Yamashita, K. / Stanfield, J.K. / Kasai, C. / Ariyasu, S. / Shoji, O. / Sugimoto, H. / Kubo, M. | |||||||||||||||
| Funding support |  Japan, 4items Japan, 4items
| |||||||||||||||
 Citation Citation |  Journal: Commun Chem / Year: 2025 Journal: Commun Chem / Year: 2025Title: XFEL crystallography reveals catalytic cycle dynamics during non-native substrate oxidation by cytochrome P450BM3. Authors: Nagao, S. / Kuwano, W. / Tosha, T. / Yamashita, K. / Stanfield, J.K. / Kasai, C. / Ariyasu, S. / Hirata, K. / Ueno, G. / Murakami, H. / Ago, H. / Yamamoto, M. / Shoji, O. / Sugimoto, H. / Kubo, M. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8yaz.cif.gz 8yaz.cif.gz | 211.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8yaz.ent.gz pdb8yaz.ent.gz | 165.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8yaz.json.gz 8yaz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8yaz_validation.pdf.gz 8yaz_validation.pdf.gz | 2.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8yaz_full_validation.pdf.gz 8yaz_full_validation.pdf.gz | 2.1 MB | Display | |
| Data in XML |  8yaz_validation.xml.gz 8yaz_validation.xml.gz | 44.9 KB | Display | |
| Data in CIF |  8yaz_validation.cif.gz 8yaz_validation.cif.gz | 61 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ya/8yaz https://data.pdbj.org/pub/pdb/validation_reports/ya/8yaz ftp://data.pdbj.org/pub/pdb/validation_reports/ya/8yaz ftp://data.pdbj.org/pub/pdb/validation_reports/ya/8yaz | HTTPS FTP |
-Related structure data
| Related structure data |  8yayC  8yb0C  8yb1C  8yb2C  8yb3C  9kt5C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 52084.340 Da / Num. of mol.: 2 / Mutation: F87A,F393H Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria)Strain: NBRC 15308 = ATCC 14581 / Production host:  References: UniProt: P14779, unspecific monooxygenase, NADPH-hemoprotein reductase |
|---|
-Non-polymers , 5 types, 509 molecules 
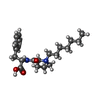







| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.71 Å3/Da / Density % sol: 54.55 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: batch mode / pH: 7.4 Details: 50 mM Tris-HCl buffer, 120 mM MgCl2, 16-18% PEG 8000, 200 uM N-Enanthyl-L-Prolyl-L-Phenylalanine, 1%(v/v) styrene |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: Y |
|---|---|
| Diffraction source | Source:  FREE ELECTRON LASER / Site: FREE ELECTRON LASER / Site:  SACLA SACLA  / Beamline: BL2 / Wavelength: 0.9539 Å / Beamline: BL2 / Wavelength: 0.9539 Å |
| Detector | Type: RAYONIX MX300-HS / Detector: CCD / Date: Dec 9, 2022 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9539 Å / Relative weight: 1 |
| Reflection | Resolution: 1.85→10 Å / Num. obs: 96618 / % possible obs: 100 % / Redundancy: 92.2 % / CC1/2: 0.972 / Net I/σ(I): 6.53 |
| Reflection shell | Resolution: 1.85→1.87 Å / Redundancy: 34.1 % / Mean I/σ(I) obs: 1.95 / Num. unique obs: 4723 / CC1/2: 0.608 |
| Serial crystallography measurement | Collection time total: 4.1 hours / Focal spot size: 19.313 µm2 / Pulse duration: 10 fsec. / Pulse photon energy: 13 keV / XFEL pulse repetition rate: 30 Hz |
| Serial crystallography sample delivery | Description: Fixed-target / Method: fixed target |
| Serial crystallography sample delivery fixed target | Crystals per unit: 50 Description: microcrystals were mounted on mesh loop and flash-frozen by liquid nitrogen Motion control: stepper motors / Sample dehydration prevention: none / Sample holding: polyimide film Sample solvent: 125 mM Tris-HCl (pH 8.3), 14% w/v PEG 8000, 60 mM MgCl2, saturated styrene, and 30% glycerol Sample unit size: 36 µm / Support base: SPINE magnet base |
| Serial crystallography data reduction | Crystal hits: 18419 / Frames indexed: 11660 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.85→9.98 Å / SU ML: 0.21 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 22.69 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.85→9.98 Å / SU ML: 0.21 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 22.69 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.85→9.98 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj






