[English] 日本語
 Yorodumi
Yorodumi- PDB-8wsv: Pre-binding structure of HosA transcriptional regulator from ente... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8wsv | ||||||
|---|---|---|---|---|---|---|---|
| Title | Pre-binding structure of HosA transcriptional regulator from enteropathogenic Escherichia coli O127:H6 (strain E2348/69) in presence of sub optimal concentration of 4-hydroxy benzoic acid | ||||||
 Components Components | Transcriptional regulator HosA | ||||||
 Keywords Keywords | DNA BINDING PROTEIN / Antibiotic resistance / MarR transcription factor / HosA / Enteropathogenic Escherichia coli | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.01 Å MOLECULAR REPLACEMENT / Resolution: 2.01 Å | ||||||
 Authors Authors | Goswami, A. / Raju, R. / Kasarla, M. / Ullah, S. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Pre-binding structure of HosA transcriptional regulator from enteropathogenic Escherichia coli O127:H6 (strain E2348/69) in presence of sub optimal concentration of 4-hydroxy benzoic acid Authors: Arpita, G. / Rukmini, R. / Mallesham, K. / Samee, U. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8wsv.cif.gz 8wsv.cif.gz | 45 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8wsv.ent.gz pdb8wsv.ent.gz | 29.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8wsv.json.gz 8wsv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8wsv_validation.pdf.gz 8wsv_validation.pdf.gz | 722.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8wsv_full_validation.pdf.gz 8wsv_full_validation.pdf.gz | 722.4 KB | Display | |
| Data in XML |  8wsv_validation.xml.gz 8wsv_validation.xml.gz | 8.4 KB | Display | |
| Data in CIF |  8wsv_validation.cif.gz 8wsv_validation.cif.gz | 11.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ws/8wsv https://data.pdbj.org/pub/pdb/validation_reports/ws/8wsv ftp://data.pdbj.org/pub/pdb/validation_reports/ws/8wsv ftp://data.pdbj.org/pub/pdb/validation_reports/ws/8wsv | HTTPS FTP |
-Related structure data
| Related structure data |  8xb7C  8pq4S  8kg0 S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 16592.092 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: HosA protein Source: (gene. exp.)  Gene: hosA / Production host:  |
|---|---|
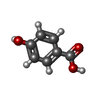
| #2: Chemical | ChemComp-PHB / |
| #3: Water | ChemComp-HOH / |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.52 Å3/Da / Density % sol: 65.06 % |
|---|---|
| Crystal grow | Temperature: 277.15 K / Method: microbatch / pH: 6.2 Details: Sodium phosphate dibasic/ Potassium phosphate monobasic, Sodium chloride, PEG 200 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.54187 Å ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.54187 Å |
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: Aug 8, 2023 / Details: Tube |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54187 Å / Relative weight: 1 |
| Reflection | Resolution: 2.01→67.47 Å / Num. obs: 14952 / % possible obs: 98.1 % / Redundancy: 7 % / Biso Wilson estimate: 13.28 Å2 / CC1/2: 0.997 / Rmerge(I) obs: 0.065 / Rpim(I) all: 0.026 / Rrim(I) all: 0.07 / Net I/σ(I): 21 |
| Reflection shell | Resolution: 2.01→2.12 Å / Redundancy: 7 % / Rmerge(I) obs: 0.116 / Mean I/σ(I) obs: 13 / Num. unique obs: 2096 / CC1/2: 0.992 / Rpim(I) all: 0.046 / Rrim(I) all: 0.125 / % possible all: 96.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 8PQ4 Resolution: 2.01→42.65 Å / SU ML: 0.21 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 23.85 / Stereochemistry target values: ML Details: TLS refinement along with occupancy was carried out for few steps. Once the partial density of the ligand was clear which aligned perfectly with ligand present in 8AGA, PHB was modeled with ...Details: TLS refinement along with occupancy was carried out for few steps. Once the partial density of the ligand was clear which aligned perfectly with ligand present in 8AGA, PHB was modeled with partial occupancy and the structure was refined using 8AGA as reference model and TLS = off.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 16.45 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.01→42.65 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj