+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 8v4j | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | |||||||||
 要素 要素 | Phosphoheptose isomerase | |||||||||
 キーワード キーワード | ISOMERASE/INHIBITOR / heptose biosynthesis / ISOMERASE-INHIBITOR complex | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報D-sedoheptulose-7-phosphate isomerase / D-sedoheptulose 7-phosphate isomerase activity / D-glycero-D-manno-heptose 7-phosphate biosynthetic process / capsule polysaccharide biosynthetic process / carbohydrate derivative binding / zinc ion binding / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Burkholderia pseudomallei 1106a (類鼻疽菌) Burkholderia pseudomallei 1106a (類鼻疽菌) | |||||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 1.31 Å 分子置換 / 解像度: 1.31 Å | |||||||||
 データ登録者 データ登録者 | Junop, M.S. / Brown, C. / Szabla, R. | |||||||||
| 資金援助 |  カナダ, 2件 カナダ, 2件
| |||||||||
 引用 引用 |  ジャーナル: J.Med.Chem. / 年: 2024 ジャーナル: J.Med.Chem. / 年: 2024タイトル: Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria. 著者: Moreau, F. / Atamanyuk, D. / Blaukopf, M. / Barath, M. / Herczeg, M. / Xavier, N.M. / Monbrun, J. / Airiau, E. / Henryon, V. / Leroy, F. / Floquet, S. / Bonnard, D. / Szabla, R. / Brown, C. / ...著者: Moreau, F. / Atamanyuk, D. / Blaukopf, M. / Barath, M. / Herczeg, M. / Xavier, N.M. / Monbrun, J. / Airiau, E. / Henryon, V. / Leroy, F. / Floquet, S. / Bonnard, D. / Szabla, R. / Brown, C. / Junop, M.S. / Kosma, P. / Gerusz, V. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  8v4j.cif.gz 8v4j.cif.gz | 96.1 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb8v4j.ent.gz pdb8v4j.ent.gz | 65 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  8v4j.json.gz 8v4j.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  8v4j_validation.pdf.gz 8v4j_validation.pdf.gz | 1.3 MB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  8v4j_full_validation.pdf.gz 8v4j_full_validation.pdf.gz | 1.3 MB | 表示 | |
| XML形式データ |  8v4j_validation.xml.gz 8v4j_validation.xml.gz | 11.9 KB | 表示 | |
| CIF形式データ |  8v4j_validation.cif.gz 8v4j_validation.cif.gz | 17.8 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/v4/8v4j https://data.pdbj.org/pub/pdb/validation_reports/v4/8v4j ftp://data.pdbj.org/pub/pdb/validation_reports/v4/8v4j ftp://data.pdbj.org/pub/pdb/validation_reports/v4/8v4j | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8v2tC C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
| 実験データセット #1 | データ参照:  10.5281/zenodo.10222807 / データの種類: diffraction image data 10.5281/zenodo.10222807 / データの種類: diffraction image data |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 単位格子 |
| ||||||||||||
| Components on special symmetry positions |
|
- 要素
要素
-タンパク質 , 1種, 1分子 A
| #1: タンパク質 | 分子量: 23393.502 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  Burkholderia pseudomallei 1106a (類鼻疽菌) Burkholderia pseudomallei 1106a (類鼻疽菌)株: K96423 / 遺伝子: gmhA / プラスミド: pNIC28-Bsa4 詳細 (発現宿主): gmhA gene cloned into pNIC28-Bsa4 plasmid backbone 発現宿主:  |
|---|
-非ポリマー , 5種, 258分子 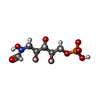








| #2: 化合物 | ChemComp-J7D / |
|---|---|
| #3: 化合物 | ChemComp-ZN / |
| #4: 化合物 | ChemComp-CL / |
| #5: 化合物 | ChemComp-NA / |
| #6: 水 | ChemComp-HOH / |
-詳細
| 研究の焦点であるリガンドがあるか | Y |
|---|
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 1.74 Å3/Da / 溶媒含有率: 33.01 % |
|---|---|
| 結晶化 | 温度: 293 K / 手法: 蒸気拡散法, ハンギングドロップ法 / pH: 4.6 詳細: 1 uL protein solution (10 mg/mL GmhA, 500 mM sodium chloride, 10 mM HEPES, pH 7.0) + 1 uL crystallization solution (10% w/v PEG4000, 0.1 M sodium acetate, pH 4.6) were mixed and suspended ...詳細: 1 uL protein solution (10 mg/mL GmhA, 500 mM sodium chloride, 10 mM HEPES, pH 7.0) + 1 uL crystallization solution (10% w/v PEG4000, 0.1 M sodium acetate, pH 4.6) were mixed and suspended over 1.5 M ammonium sulfate. Once crystals were grown, 0.2 uL ligand solution (50 mM Mut148233, 50 mM HEPES, pH 7.5) was added to the drops and allowed to soak for 90 minutes. Temp details: Incubator |
-データ収集
| 回折 | 平均測定温度: 100 K / Ambient temp details: Nitrogen cryo-stream / Serial crystal experiment: N |
|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  NSLS NSLS  / ビームライン: X29A / 波長: 1.075 Å / ビームライン: X29A / 波長: 1.075 Å |
| 検出器 | タイプ: ADSC QUANTUM 315 / 検出器: CCD / 日付: 2013年6月10日 |
| 放射 | プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray |
| 放射波長 | 波長: 1.075 Å / 相対比: 1 |
| 反射 | 解像度: 1.31→36.83 Å / Num. obs: 42503 / % possible obs: 99.68 % / 冗長度: 13.7 % / Biso Wilson estimate: 10.67 Å2 / CC1/2: 1 / CC star: 1 / Rmerge(I) obs: 0.03807 / Rpim(I) all: 0.01072 / Rrim(I) all: 0.03959 / Net I/σ(I): 41.13 |
| 反射 シェル | 解像度: 1.31→1.357 Å / 冗長度: 13.7 % / Rmerge(I) obs: 0.2714 / Mean I/σ(I) obs: 8.89 / Num. unique obs: 4190 / CC1/2: 0.985 / CC star: 0.996 / Rpim(I) all: 0.07575 / Rrim(I) all: 0.2819 / % possible all: 100 |
- 解析
解析
| ソフトウェア |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法:  分子置換 / 解像度: 1.31→36.83 Å / SU ML: 0.0963 / 交差検証法: FREE R-VALUE / σ(F): 1.34 / 位相誤差: 16.2308 分子置換 / 解像度: 1.31→36.83 Å / SU ML: 0.0963 / 交差検証法: FREE R-VALUE / σ(F): 1.34 / 位相誤差: 16.2308 立体化学のターゲット値: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | 減衰半径: 0.9 Å / VDWプローブ半径: 1.11 Å / 溶媒モデル: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso mean: 15.14 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: LAST / 解像度: 1.31→36.83 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS精密化 シェル |
|
 ムービー
ムービー コントローラー
コントローラー



 PDBj
PDBj



