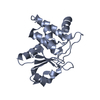
Entry Database : PDB / ID : 8tk5Title HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with HEPES Dual specificity protein phosphatase 3 Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 1.9 Å Authors Aleshin, A.E. / Wu, J. / Lambert, L.J. / Cosford, N.D.P. / Tautz, L. Funding support Organization Grant number Country National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) 1R21 AI160161
Journal : Acs Omega / Year : 2025Title : Fragment Screening Identifies Novel Allosteric Binders and Binding Sites in the VHR ( DUSP3 ) Phosphatase.Authors : Wu, J. / Baranowski, M.R. / Aleshin, A.E. / Isiorho, E.A. / Lambert, L.J. / De Backer, L.J.S. / Han, Y.N. / Das, R. / Sheffler, D.J. / Bobkov, A.A. / Lemberikman, A.M. / Keedy, D.A. / Cosford, N.D.P. / Tautz, L. History Deposition Jul 25, 2023 Deposition site / Processing site Revision 1.0 Aug 14, 2024 Provider / Type Revision 1.1 Feb 26, 2025 Group / Structure summary / Category / citation_author / pdbx_entry_detailsItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _pdbx_entry_details.has_protein_modification
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å
MOLECULAR REPLACEMENT / Resolution: 1.9 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Acs Omega / Year: 2025
Journal: Acs Omega / Year: 2025 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 8tk5.cif.gz
8tk5.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb8tk5.ent.gz
pdb8tk5.ent.gz PDB format
PDB format 8tk5.json.gz
8tk5.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/tk/8tk5
https://data.pdbj.org/pub/pdb/validation_reports/tk/8tk5 ftp://data.pdbj.org/pub/pdb/validation_reports/tk/8tk5
ftp://data.pdbj.org/pub/pdb/validation_reports/tk/8tk5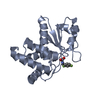



 F&H Search
F&H Search Links
Links Assembly
Assembly


 Components
Components Homo sapiens (human) / Gene: DUSP3, VHR / Production host:
Homo sapiens (human) / Gene: DUSP3, VHR / Production host: 









 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRL
SSRL  / Beamline: BL12-2 / Wavelength: 0.97946 Å
/ Beamline: BL12-2 / Wavelength: 0.97946 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 1.9→38.2 Å / Cor.coef. Fo:Fc: 0.968 / Cor.coef. Fo:Fc free: 0.939 / SU B: 2.855 / SU ML: 0.084 / Cross valid method: THROUGHOUT / ESU R: 0.146 / ESU R Free: 0.133 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MOLECULAR REPLACEMENT / Resolution: 1.9→38.2 Å / Cor.coef. Fo:Fc: 0.968 / Cor.coef. Fo:Fc free: 0.939 / SU B: 2.855 / SU ML: 0.084 / Cross valid method: THROUGHOUT / ESU R: 0.146 / ESU R Free: 0.133 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller


 PDBj
PDBj





