+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8qx6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Novel laminarin-binding CBM X584 | ||||||
 Components Components | PKD domain-containing protein | ||||||
 Keywords Keywords | SUGAR BINDING PROTEIN / CBM X584 / Surface glycan binding protein / carbohydrate binding module / ligand complex | ||||||
| Function / homology | Polycystic kidney disease (PKD) domain profile. / PKD domain / Prokaryotic membrane lipoprotein lipid attachment site profile. / PKD domain-containing protein Function and homology information Function and homology information | ||||||
| Biological species |  Christiangramia forsetii KT0803 (bacteria) Christiangramia forsetii KT0803 (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Zuehlke, M.K. / Jeudy, A. / Czjzek, M. | ||||||
| Funding support |  Germany, 1items Germany, 1items
| ||||||
 Citation Citation |  Journal: Environ.Microbiol. / Year: 2024 Journal: Environ.Microbiol. / Year: 2024Title: Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms. Authors: Zuhlke, M.K. / Ficko-Blean, E. / Bartosik, D. / Terrapon, N. / Jeudy, A. / Jam, M. / Wang, F. / Welsch, N. / Durwald, A. / Martin, L.T. / Larocque, R. / Jouanneau, D. / Eisenack, T. / ...Authors: Zuhlke, M.K. / Ficko-Blean, E. / Bartosik, D. / Terrapon, N. / Jeudy, A. / Jam, M. / Wang, F. / Welsch, N. / Durwald, A. / Martin, L.T. / Larocque, R. / Jouanneau, D. / Eisenack, T. / Thomas, F. / Trautwein-Schult, A. / Teeling, H. / Becher, D. / Schweder, T. / Czjzek, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8qx6.cif.gz 8qx6.cif.gz | 127.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8qx6.ent.gz pdb8qx6.ent.gz | 101.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8qx6.json.gz 8qx6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qx/8qx6 https://data.pdbj.org/pub/pdb/validation_reports/qx/8qx6 ftp://data.pdbj.org/pub/pdb/validation_reports/qx/8qx6 ftp://data.pdbj.org/pub/pdb/validation_reports/qx/8qx6 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 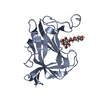
| ||||||||
| 2 | 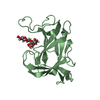
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 18092.477 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: isolated carbohydrate binding module part of the protein with Uniprot code A0M709 Source: (gene. exp.)  Christiangramia forsetii KT0803 (bacteria) Christiangramia forsetii KT0803 (bacteria)Gene: GFO_3465 Production host:  References: UniProt: A0M709 #2: Polysaccharide | Source method: isolated from a genetically manipulated source #3: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 38 % |
|---|---|
| Crystal grow | Temperature: 292 K / Method: counter-diffusion / pH: 7 Details: 2.5 M ammonium sulfate, 0.1 M BIS-TRIS propane pH 7.0, 6% ethanol and 4% polyethylenglycol (PEG) 6000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: N2 gasstream / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.98 Å / Beamline: PROXIMA 1 / Wavelength: 0.98 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Mar 23, 2023 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→47.42 Å / Num. obs: 23977 / % possible obs: 99.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 12.9 % / CC1/2: 0.99 / Rmerge(I) obs: 0.148 / Rpim(I) all: 0.043 / Net I/σ(I): 10.9 |
| Reflection shell | Resolution: 1.9→1.95 Å / Redundancy: 10.2 % / Rmerge(I) obs: 1.81 / Mean I/σ(I) obs: 1.3 / Num. unique obs: 1610 / CC1/2: 0.58 / Rpim(I) all: 0.59 / % possible all: 96.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.9→47.42 Å / SU ML: 0.153 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.213 / ESU R Free: 0.186 MOLECULAR REPLACEMENT / Resolution: 1.9→47.42 Å / SU ML: 0.153 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.213 / ESU R Free: 0.186
| ||||||||||||||||||||
| Displacement parameters | Biso mean: 32.27 Å2 | ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→47.42 Å
| ||||||||||||||||||||
| Refine LS restraints | Type: NCS Local / Dev ideal: 0.14 / Weight: 0.05 | ||||||||||||||||||||
| LS refinement shell | Resolution: 1.9→1.95 Å
|
 Movie
Movie Controller
Controller



 PDBj
PDBj





