[English] 日本語
 Yorodumi
Yorodumi- PDB-8p8g: Nitrogenase MoFe protein from A. vinelandii beta double mutant D3... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8p8g | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nitrogenase MoFe protein from A. vinelandii beta double mutant D353G/D357G | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | OXIDOREDUCTASE / Molybdenum nitrogenase / N2-fixation / FeMocofactor / metal binding site / P cluster / NH3 production | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / iron-sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.55 Å MOLECULAR REPLACEMENT / Resolution: 1.55 Å | ||||||||||||
 Authors Authors | Maslac, N. / Wagner, T. | ||||||||||||
| Funding support |  Germany, Germany,  Switzerland, 3items Switzerland, 3items
| ||||||||||||
 Citation Citation |  Journal: Jacs Au / Year: 2023 Journal: Jacs Au / Year: 2023Title: The Mononuclear Metal-Binding Site of Mo-Nitrogenase Is Not Required for Activity. Authors: Cadoux, C. / Maslac, N. / Di Luzio, L. / Ratcliff, D. / Gu, W. / Wagner, T. / Milton, R.D. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8p8g.cif.gz 8p8g.cif.gz | 847.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8p8g.ent.gz pdb8p8g.ent.gz | 690.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8p8g.json.gz 8p8g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/p8/8p8g https://data.pdbj.org/pub/pdb/validation_reports/p8/8p8g ftp://data.pdbj.org/pub/pdb/validation_reports/p8/8p8g ftp://data.pdbj.org/pub/pdb/validation_reports/p8/8p8g | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|---|
| Experimental dataset #1 | Data reference:  10.5281/zenodo.7973626 / Data set type: diffraction image data / Metadata reference: 10.5281/zenodo.7973626 10.5281/zenodo.7973626 / Data set type: diffraction image data / Metadata reference: 10.5281/zenodo.7973626 |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 2 types, 4 molecules ACBD
| #1: Protein | Mass: 56467.234 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azotobacter vinelandii DJ (bacteria) / Strain: DJ / Tissue: / / Cell line: / / Gene: nifD / Organ: / Azotobacter vinelandii DJ (bacteria) / Strain: DJ / Tissue: / / Cell line: / / Gene: nifD / Organ: /Plasmid details: Produces the MoFe protein with a poly(histidine)8 tag on the N-term of NifD and a double substituted NifK mutant Production host:  Azotobacter vinelandii DJ (bacteria) / References: UniProt: C1DGZ7 Azotobacter vinelandii DJ (bacteria) / References: UniProt: C1DGZ7#2: Protein | Mass: 59419.809 Da / Num. of mol.: 2 / Mutation: D353G and D357G Source method: isolated from a genetically manipulated source Details: The double substitution D353G and D357G was introduced. Source: (gene. exp.)  Azotobacter vinelandii DJ (bacteria) / Strain: DJ / Tissue: / / Cell line: / / Gene: nifK, Avin_01400 / Organ: / Azotobacter vinelandii DJ (bacteria) / Strain: DJ / Tissue: / / Cell line: / / Gene: nifK, Avin_01400 / Organ: /Plasmid details: Produces the MoFe protein with a poly(histidine)8 tag on the N-term of NifD and a double substituted NifK mutant Production host:  Azotobacter vinelandii DJ (bacteria) / References: UniProt: C1DGZ8, nitrogenase Azotobacter vinelandii DJ (bacteria) / References: UniProt: C1DGZ8, nitrogenase |
|---|
-Non-polymers , 10 types, 2222 molecules 

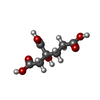
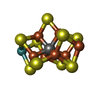








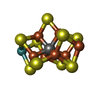






| #3: Chemical | ChemComp-EDO / #4: Chemical | #5: Chemical | #6: Chemical | #7: Chemical | #8: Chemical | ChemComp-GOL / | #9: Chemical | #10: Chemical | #11: Chemical | ChemComp-NA / | #12: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.74 Å3/Da / Density % sol: 55.06 % Description: Brown plate, which appeared in a couple of weeks |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, sitting drop / pH: 5.6 Details: The beta-D353G/D357G MoFe-protein was crystallized anaerobically at 17.5 mg/mL under 100% N2 atmosphere. The protein was spotted as a sitting drop to 96-Well MRC 2-Drop polystyrene ...Details: The beta-D353G/D357G MoFe-protein was crystallized anaerobically at 17.5 mg/mL under 100% N2 atmosphere. The protein was spotted as a sitting drop to 96-Well MRC 2-Drop polystyrene Crystallization Plates (SWISSCI) containing 90 uL of crystallization solution in the reservoir. Each drop contained 0.5 uL of protein sample and 0.5 uL of crystallization solution. Crystals were obtained in the crystallization solution containing 10 % w/v Polyethylene glycol 10,000; 2 % v/v 1,4-Dioxane; 100 mM tri-Sodium citrate; pH 5.6, and 1 mM polyoxotungstate [TeW6O24]6- (TEW). Sealed plates were stored inside a Coy anaerobic chamber filled with an atmosphere of N2:H2 97:3 at 20 C. Crystals were soaked in the crystallization solution supplemented with 30% v/v ethylene glycol for a few seconds before freezing in liquid nitrogen. PH range: / / Temp details: / |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1.00002 Å / Beamline: X06DA / Wavelength: 1.00002 Å |
| Detector | Type: DECTRIS PILATUS 2M-F / Detector: PIXEL / Date: Oct 16, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.00002 Å / Relative weight: 1 |
| Reflection | Resolution: 1.55→101.46 Å / Num. obs: 176043 / % possible obs: 94 % / Redundancy: 8.1 % / Biso Wilson estimate: 14 Å2 / CC1/2: 0.995 / Rmerge(I) obs: 0.177 / Rpim(I) all: 0.066 / Rrim(I) all: 0.189 / Net I/σ(I): 8.1 |
| Reflection shell | Resolution: 1.55→1.76 Å / Redundancy: 9.3 % / Rmerge(I) obs: 1.89 / Mean I/σ(I) obs: 1.8 / Num. unique obs: 8391 / CC1/2: 0.566 / Rpim(I) all: 0.652 / Rrim(I) all: 2.001 / % possible all: 80.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.55→48.32 Å / SU ML: 0.14 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 22.28 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.55→48.32 Å / SU ML: 0.14 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 22.28 / Stereochemistry target values: MLDetails: The model was manually rebuilt with COOT and further refined with PHENIX. During the refinement, a translational-libration screw was applied with the model containing hydrogens added in the ...Details: The model was manually rebuilt with COOT and further refined with PHENIX. During the refinement, a translational-libration screw was applied with the model containing hydrogens added in the riding position during the last refinement cycles. Hydrogens were removed in the final deposited model.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 18.62 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.55→48.32 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj








