Entry Database : PDB / ID : 8h5vTitle Crystal structure of the FleQ domain of Vibrio cholerae FlrA flagellar regulatory protein A Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Vibrio cholerae O395 (bacteria)Method / / / Resolution : 2 Å Authors Dasgupta, J. / Chakraborty, S. Funding support Organization Grant number Country Department of Science & Technology (DST, India)
Journal : Febs Lett. / Year : 2023Title : The N-terminal FleQ domain of the Vibrio cholerae flagellar master regulator FlrA plays pivotal structural roles in stabilizing its active state.Authors : Chakraborty, S. / Agarwal, S. / Bakshi, A. / Dey, S. / Biswas, M. / Ghosh, B. / Dasgupta, J. History Deposition Oct 14, 2022 Deposition site / Processing site Revision 1.0 Nov 2, 2022 Provider / Type Revision 1.1 May 15, 2024 Group / Database referencesCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / citation / citation_author Item _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Vibrio cholerae O395 (bacteria)
Vibrio cholerae O395 (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å
MOLECULAR REPLACEMENT / Resolution: 2 Å  Authors
Authors India, 1items
India, 1items  Citation
Citation Journal: Febs Lett. / Year: 2023
Journal: Febs Lett. / Year: 2023 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 8h5v.cif.gz
8h5v.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb8h5v.ent.gz
pdb8h5v.ent.gz PDB format
PDB format 8h5v.json.gz
8h5v.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/h5/8h5v
https://data.pdbj.org/pub/pdb/validation_reports/h5/8h5v ftp://data.pdbj.org/pub/pdb/validation_reports/h5/8h5v
ftp://data.pdbj.org/pub/pdb/validation_reports/h5/8h5v F&H Search
F&H Search Links
Links Assembly
Assembly



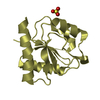
 Components
Components Vibrio cholerae O395 (bacteria) / Gene: EYB64_02180 / Production host:
Vibrio cholerae O395 (bacteria) / Gene: EYB64_02180 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  RRCAT INDUS-2
RRCAT INDUS-2  / Beamline: PX-BL21 / Wavelength: 0.97893 Å
/ Beamline: PX-BL21 / Wavelength: 0.97893 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller



 PDBj
PDBj





