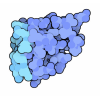
| Entry | Database: PDB / ID: 8h2c
|
|---|
| Title | Crystal structure of the pseudaminic acid synthase PseI from Campylobacter jejuni |
|---|
 Components Components | Pseudaminic acid synthase |
|---|
 Keywords Keywords | BIOSYNTHETIC PROTEIN / Pseudaminic acid synthase / TIM barrel fold / Type III antifreeze-like domain / condensation reaction |
|---|
| Function / homology |  Function and homology information Function and homology information
Pseudaminic acid biosynthesis, PseI / N-acetylneuraminic acid synthase, N-terminal / : / NeuB family / SAF domain / SAF / SAF domain / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal / Antifreeze protein-like domain profile. / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal domain superfamily / Aldolase-type TIM barrelSimilarity search - Domain/homology |
|---|
| Biological species |   Campylobacter jejuni (Campylobacter) Campylobacter jejuni (Campylobacter) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å |
|---|
 Authors Authors | Song, W.S. / Park, M.A. / Ki, D.U. / Yoon, S.I. |
|---|
| Funding support |  Korea, Republic Of, 2items Korea, Republic Of, 2items | Organization | Grant number | Country |
|---|
| National Research Foundation (NRF, Korea) | 2019R1A2C1002100 |  Korea, Republic Of Korea, Republic Of | | National Research Foundation (NRF, Korea) | 2019R1I1A1A01047141 |  Korea, Republic Of Korea, Republic Of |
|
|---|
 Citation Citation |  Journal: Biochem.Biophys.Res.Commun. / Year: 2022 Journal: Biochem.Biophys.Res.Commun. / Year: 2022
Title: Structural analysis of the pseudaminic acid synthase PseI from Campylobacter jejuni.
Authors: Song, W.S. / Park, M.A. / Ki, D.U. / Yoon, S.I. |
|---|
| History | | Deposition | Oct 5, 2022 | Deposition site: PDBJ / Processing site: RCSB |
|---|
| Revision 1.0 | Nov 9, 2022 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Apr 3, 2024 | Group: Data collection / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / pdbx_initial_refinement_model / struct_ncs_dom_lim
Item: _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id ..._struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å
MOLECULAR REPLACEMENT / Resolution: 2.9 Å  Authors
Authors Korea, Republic Of, 2items
Korea, Republic Of, 2items  Citation
Citation Journal: Biochem.Biophys.Res.Commun. / Year: 2022
Journal: Biochem.Biophys.Res.Commun. / Year: 2022 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 8h2c.cif.gz
8h2c.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb8h2c.ent.gz
pdb8h2c.ent.gz PDB format
PDB format 8h2c.json.gz
8h2c.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 8h2c_validation.pdf.gz
8h2c_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 8h2c_full_validation.pdf.gz
8h2c_full_validation.pdf.gz 8h2c_validation.xml.gz
8h2c_validation.xml.gz 8h2c_validation.cif.gz
8h2c_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/h2/8h2c
https://data.pdbj.org/pub/pdb/validation_reports/h2/8h2c ftp://data.pdbj.org/pub/pdb/validation_reports/h2/8h2c
ftp://data.pdbj.org/pub/pdb/validation_reports/h2/8h2c F&H Search
F&H Search Links
Links Assembly
Assembly


 Movie
Movie Controller
Controller


 PDBj
PDBj