| ソフトウェア | | 名称 | バージョン | 分類 |
|---|
| PHENIX | 1.20.1_4487| 精密化 | | XDS | | データ削減 | | XDS | | データスケーリング | | PHENIX | | 位相決定 | |
|
|---|
| 精密化 | 構造決定の手法:  分子置換 / 解像度: 1.78→36.37 Å / SU ML: 0.21 / 交差検証法: FREE R-VALUE / σ(F): 1.97 / 位相誤差: 20.64 / 立体化学のターゲット値: ML 分子置換 / 解像度: 1.78→36.37 Å / SU ML: 0.21 / 交差検証法: FREE R-VALUE / σ(F): 1.97 / 位相誤差: 20.64 / 立体化学のターゲット値: ML
| Rfactor | 反射数 | %反射 |
|---|
| Rfree | 0.1944 | 2000 | 7.26 % |
|---|
| Rwork | 0.1613 | - | - |
|---|
| obs | 0.1637 | 27533 | 93.95 % |
|---|
|
|---|
| 溶媒の処理 | 減衰半径: 0.9 Å / VDWプローブ半径: 1.1 Å / 溶媒モデル: FLAT BULK SOLVENT MODEL |
|---|
| 精密化ステップ | サイクル: LAST / 解像度: 1.78→36.37 Å
| タンパク質 | 核酸 | リガンド | 溶媒 | 全体 |
|---|
| 原子数 | 2096 | 0 | 26 | 374 | 2496 |
|---|
|
|---|
| 拘束条件 | | Refine-ID | タイプ | Dev ideal | 数 |
|---|
| X-RAY DIFFRACTION | f_bond_d| 0.014 | 2173 | | X-RAY DIFFRACTION | f_angle_d| 1.283 | 2951 | | X-RAY DIFFRACTION | f_dihedral_angle_d| 19.177 | 301 | | X-RAY DIFFRACTION | f_chiral_restr| 0.079 | 335 | | X-RAY DIFFRACTION | f_plane_restr| 0.012 | 374 | | | | | |
|
|---|
| LS精密化 シェル | | 解像度 (Å) | Rfactor Rfree | Num. reflection Rfree | Rfactor Rwork | Num. reflection Rwork | Refine-ID | % reflection obs (%) |
|---|
| 1.78-1.83 | 0.3218 | 134 | 0.2468 | 1707 | X-RAY DIFFRACTION | 88 | | 1.83-1.88 | 0.2237 | 130 | 0.2318 | 1666 | X-RAY DIFFRACTION | 86 | | 1.88-1.93 | 0.2587 | 145 | 0.1984 | 1848 | X-RAY DIFFRACTION | 94 | | 1.93-1.99 | 0.2399 | 141 | 0.1981 | 1801 | X-RAY DIFFRACTION | 94 | | 1.99-2.06 | 0.2452 | 147 | 0.1735 | 1867 | X-RAY DIFFRACTION | 95 | | 2.06-2.15 | 0.2083 | 144 | 0.1615 | 1847 | X-RAY DIFFRACTION | 95 | | 2.15-2.24 | 0.1896 | 144 | 0.1648 | 1839 | X-RAY DIFFRACTION | 95 | | 2.24-2.36 | 0.208 | 144 | 0.1617 | 1832 | X-RAY DIFFRACTION | 94 | | 2.36-2.51 | 0.2173 | 145 | 0.1692 | 1856 | X-RAY DIFFRACTION | 95 | | 2.51-2.7 | 0.2157 | 138 | 0.1726 | 1762 | X-RAY DIFFRACTION | 92 | | 2.7-2.98 | 0.195 | 149 | 0.1635 | 1914 | X-RAY DIFFRACTION | 97 | | 2.98-3.41 | 0.1927 | 147 | 0.1513 | 1867 | X-RAY DIFFRACTION | 98 | | 3.41-4.29 | 0.1588 | 146 | 0.1298 | 1870 | X-RAY DIFFRACTION | 96 | | 4.29-36.37 | 0.1535 | 146 | 0.1537 | 1857 | X-RAY DIFFRACTION | 96 |
|
|---|
| 精密化 TLS | 手法: refined / Refine-ID: X-RAY DIFFRACTION | ID | L11 (°2) | L12 (°2) | L13 (°2) | L22 (°2) | L23 (°2) | L33 (°2) | S11 (Å °) | S12 (Å °) | S13 (Å °) | S21 (Å °) | S22 (Å °) | S23 (Å °) | S31 (Å °) | S32 (Å °) | S33 (Å °) | T11 (Å2) | T12 (Å2) | T13 (Å2) | T22 (Å2) | T23 (Å2) | T33 (Å2) | Origin x (Å) | Origin y (Å) | Origin z (Å) |
|---|
| 1 | 1.8014 | 0.104 | -0.4247 | 5.1891 | -2.0358 | 3.7833 | -0.0997 | -0.1072 | -0.2528 | 0.5109 | -0.3066 | -0.3908 | -0.0518 | 0.1473 | 0.3322 | 0.1802 | -0.0233 | -0.0051 | 0.2353 | 0.0315 | 0.1973 | -0.4516 | -4.6256 | 7.0703 | | 2 | 2.3867 | 0.505 | -0.6243 | 2.594 | 0.5311 | 2.4468 | 0.1248 | -0.2821 | -0.0243 | 0.1017 | -0.0201 | -0.3546 | -0.0967 | 0.2209 | -0.1318 | 0.1589 | -0.0093 | -0.0024 | 0.2218 | 0.0096 | 0.2085 | 7.608 | 1.1475 | 8.6869 | | 3 | 0.7271 | 0.6103 | 0.8055 | 0.6729 | 0.2719 | 2.357 | 0.0507 | 0.4065 | -0.2044 | -0.101 | 0.0433 | 0.0388 | 0.0875 | 0.1514 | -0.0882 | 0.1981 | 0.0194 | 0.0139 | 0.2629 | -0.006 | 0.1912 | -1.8652 | -3.2368 | -5.4655 | | 4 | 0.997 | 0.4227 | 0.8079 | 1.7286 | -0.8969 | 5.8899 | -0.0004 | -0.0069 | -0.4666 | 0.0561 | 0.1814 | -0.2725 | 0.5884 | -0.4246 | -0.1439 | 0.2844 | -0.0309 | -0.0302 | 0.2347 | -0.0298 | 0.2471 | -6.8618 | -9.7225 | -3.956 | | 5 | 1.4727 | -0.6663 | 0.3813 | 1.8761 | -0.7873 | 2.9613 | 0.0749 | 0.0727 | -0.0389 | -0.0842 | -0.008 | 0.2052 | 0.0471 | -0.2883 | -0.044 | 0.1265 | -0.002 | -0.0106 | 0.1844 | 0.0207 | 0.1398 | -8.4397 | -0.0395 | 2.1128 | | 6 | 2.0162 | 0.3381 | 0.052 | 2.2345 | 0.0956 | 1.7969 | 0.1853 | -0.2192 | 0.1327 | 0.2948 | -0.1564 | 0.0282 | -0.136 | -0.0804 | -0.0401 | 0.1673 | -0.0011 | 0.01 | 0.1768 | 0.0109 | 0.1542 | -6.183 | 6.6541 | 8.4695 | | 7 | 3.2357 | 0.683 | -0.3619 | 2.063 | 0.8819 | 1.416 | 0.1172 | -0.0098 | 0.1318 | -0.0271 | -0.0631 | 0.2819 | -0.1598 | -0.1616 | 0.0291 | 0.1718 | 0.0099 | 0.0109 | 0.2369 | -0.0009 | 0.1946 | -11.0415 | 6.0385 | 9.6565 | | 8 | 3.7576 | 0.1434 | -0.558 | 2.4534 | -0.2518 | 1.7689 | 0.0511 | -0.6331 | -0.0624 | 0.3868 | -0.0044 | -0.1629 | 0.054 | 0.0283 | -0.0679 | 0.2113 | -0.0482 | -0.0162 | 0.3117 | 0.0253 | 0.1497 | -0.3255 | 2.0462 | 17.7501 | | 9 | 9.4947 | -1.1121 | -1.3196 | 2.0263 | -0.0207 | 8.7398 | 0.9273 | -0.7602 | -0.5127 | 0.2154 | 0.1557 | 1.4794 | -0.149 | -0.7251 | 0.5539 | 0.2909 | 0.0317 | -0.0882 | 0.4051 | 0.2557 | 0.5351 | -12.0196 | -7.4998 | 20.4483 | | 10 | 2.6928 | -0.9348 | -0.5955 | 1.7077 | -0.2899 | 2.3908 | 0.0505 | -0.1877 | 0.1563 | 0.084 | 0.0233 | -0.1225 | -0.0269 | 0.3251 | -0.071 | 0.1385 | -0.0011 | -0.0063 | 0.163 | -0.026 | 0.1557 | -6.179 | 17.9354 | 33.4216 | | 11 | 1.8695 | 0.3037 | -0.8912 | 1.8754 | 0.0463 | 1.5534 | 0.0676 | -0.4308 | -0.2885 | 0.235 | -0.1552 | 0.0231 | 0.5412 | 0.2674 | 0.0164 | 0.2986 | 0.026 | -0.0168 | 0.2071 | 0.044 | 0.1941 | -16.1697 | 10.7807 | 45.0332 | | 12 | 1.8756 | 0.1104 | -0.1708 | 2.1839 | 0.1768 | 2.4932 | -0.0066 | 0.0095 | -0.1112 | -0.0159 | 0.0128 | -0.145 | 0.198 | -0.0889 | 0.046 | 0.1608 | 0.0009 | -0.0111 | 0.1436 | -0.0167 | 0.1711 | -17.5376 | 12.3477 | 32.6487 | | 13 | 1.062 | 0.3219 | -0.1999 | 3.9272 | 0.0711 | 1.9942 | -0.0061 | -0.1674 | -0.2049 | 0.3277 | -0.0576 | 0.4776 | 0.1264 | -0.1621 | 0.0234 | 0.187 | -0.0094 | 0.0092 | 0.1849 | 0.005 | 0.185 | -22.9512 | 14.3027 | 40.0939 | | 14 | 1.8706 | -0.2606 | -0.8801 | 1.7479 | 0.5761 | 2.767 | 0.0267 | 0.2652 | -0.0426 | -0.1584 | -0.1063 | 0.081 | -0.1003 | -0.1991 | 0.0263 | 0.1568 | -0.0081 | -0.0026 | 0.1647 | 0.0016 | 0.1749 | -17.9375 | 15.4198 | 27.6167 | | 15 | 4.319 | -2.7084 | -0.0954 | 4.1541 | 0.2659 | 2.427 | 0.118 | 0.2953 | -0.5268 | -0.0273 | 0.0304 | 0.5156 | 0.4903 | -0.7567 | 0.0235 | 0.1239 | -0.0244 | -0.0116 | 0.2777 | 0.0153 | 0.1928 | -27.8845 | 12.7665 | 30.7836 | | 16 | 2.4122 | 0.2151 | -1.1124 | 1.087 | -0.6491 | 1.5582 | -0.0564 | 0.1998 | 0.4468 | -0.0783 | 0.0179 | 0.0978 | 0.1498 | -0.1808 | -0.0249 | 0.1589 | -0.0023 | -0.0167 | 0.2266 | 0.014 | 0.1916 | -19.3188 | 19.0942 | 24.844 | | 17 | 0.295 | 0.2283 | 0.1062 | 0.4448 | -0.1916 | 0.3753 | 0.3302 | 0.0284 | 0.7348 | -0.145 | 0.0335 | 0.0521 | -0.4717 | -0.0857 | -0.1338 | 0.2234 | -0.0034 | 0.0343 | 0.1847 | 0.0398 | 0.2951 | -13.5833 | 27.0538 | 27.8063 |
|
|---|
| 精密化 TLSグループ | | ID | Refine-ID | Refine TLS-ID | Selection details |
|---|
| 1 | X-RAY DIFFRACTION | 1 | chain 'A' and (resid 1 through 10 )| 2 | X-RAY DIFFRACTION | 2 | chain 'A' and (resid 11 through 35 )| 3 | X-RAY DIFFRACTION | 3 | chain 'A' and (resid 36 through 47 )| 4 | X-RAY DIFFRACTION | 4 | chain 'A' and (resid 48 through 61 )| 5 | X-RAY DIFFRACTION | 5 | chain 'A' and (resid 62 through 89 )| 6 | X-RAY DIFFRACTION | 6 | chain 'A' and (resid 90 through 106 )| 7 | X-RAY DIFFRACTION | 7 | chain 'A' and (resid 107 through 120 )| 8 | X-RAY DIFFRACTION | 8 | chain 'A' and (resid 121 through 129 )| 9 | X-RAY DIFFRACTION | 9 | chain 'A' and (resid 130 through 136 )| 10 | X-RAY DIFFRACTION | 10 | chain 'B' and (resid 3 through 35 )| 11 | X-RAY DIFFRACTION | 11 | chain 'B' and (resid 36 through 61 )| 12 | X-RAY DIFFRACTION | 12 | chain 'B' and (resid 62 through 77 )| 13 | X-RAY DIFFRACTION | 13 | chain 'B' and (resid 78 through 88 )| 14 | X-RAY DIFFRACTION | 14 | chain 'B' and (resid 89 through 106 )| 15 | X-RAY DIFFRACTION | 15 | chain 'B' and (resid 107 through 111 )| 16 | X-RAY DIFFRACTION | 16 | chain 'B | | | | | | | | | | | | | | | |
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 1.78 Å
分子置換 / 解像度: 1.78 Å  データ登録者
データ登録者 米国, 1件
米国, 1件  引用
引用 ジャーナル: To Be Published
ジャーナル: To Be Published 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 8fz9.cif.gz
8fz9.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb8fz9.ent.gz
pdb8fz9.ent.gz PDB形式
PDB形式 8fz9.json.gz
8fz9.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 8fz9_validation.pdf.gz
8fz9_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 8fz9_full_validation.pdf.gz
8fz9_full_validation.pdf.gz 8fz9_validation.xml.gz
8fz9_validation.xml.gz 8fz9_validation.cif.gz
8fz9_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/fz/8fz9
https://data.pdbj.org/pub/pdb/validation_reports/fz/8fz9 ftp://data.pdbj.org/pub/pdb/validation_reports/fz/8fz9
ftp://data.pdbj.org/pub/pdb/validation_reports/fz/8fz9 F&H 検索
F&H 検索 リンク
リンク 集合体
集合体
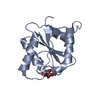

 要素
要素

 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 24-ID-E / 波長: 0.97918 Å
/ ビームライン: 24-ID-E / 波長: 0.97918 Å 解析
解析 分子置換 / 解像度: 1.78→36.37 Å / SU ML: 0.21 / 交差検証法: FREE R-VALUE / σ(F): 1.97 / 位相誤差: 20.64 / 立体化学のターゲット値: ML
分子置換 / 解像度: 1.78→36.37 Å / SU ML: 0.21 / 交差検証法: FREE R-VALUE / σ(F): 1.97 / 位相誤差: 20.64 / 立体化学のターゲット値: ML ムービー
ムービー コントローラー
コントローラー



 PDBj
PDBj


