[English] 日本語
 Yorodumi
Yorodumi- PDB-8cns: The Hybrid Cluster Protein from the thermophilic methanogen Metha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8cns | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Hybrid Cluster Protein from the thermophilic methanogen Methanothermococcus thermolithotrophicus in a mixed redox state after soaking with hydroxylamine, at 1.36-A resolution. | |||||||||
 Components Components | Hybrid cluster protein from Methanothermococcus thermolithotrophicus | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / Hybrid cluster / prismane protein / Hybrid cluster protein / methanogenic archaea / anaerobic biochemistry / thermophile / metallocluster / Nitric oxide reductase / hydroxylamine | |||||||||
| Function / homology | ACETATE ION / FORMIC ACID / HYDROXYAMINE / DI(HYDROXYETHYL)ETHER / 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE / FE4-S3 CLUSTER / IRON/SULFUR CLUSTER / hybrid cluster Function and homology information Function and homology information | |||||||||
| Biological species |  Methanothermococcus thermolithotrophicus DSM 2095 (archaea) Methanothermococcus thermolithotrophicus DSM 2095 (archaea) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.36 Å MOLECULAR REPLACEMENT / Resolution: 1.36 Å | |||||||||
 Authors Authors | Lemaire, O.N. / Wagner, T. | |||||||||
| Funding support |  Germany, 2items Germany, 2items
| |||||||||
 Citation Citation |  Journal: Front Microbiol / Year: 2023 Journal: Front Microbiol / Year: 2023Title: Structural and biochemical elucidation of class I hybrid cluster protein natively extracted from a marine methanogenic archaeon. Authors: Lemaire, O.N. / Belhamri, M. / Wagner, T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8cns.cif.gz 8cns.cif.gz | 269.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8cns.ent.gz pdb8cns.ent.gz | 213.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8cns.json.gz 8cns.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cn/8cns https://data.pdbj.org/pub/pdb/validation_reports/cn/8cns ftp://data.pdbj.org/pub/pdb/validation_reports/cn/8cns ftp://data.pdbj.org/pub/pdb/validation_reports/cn/8cns | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8cnrC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 60367.773 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: Compared to the automatic annotation, the sequence has a five-residue extension in its N-terminal (MRPSK). The cysteine 402 has a thiol addition in its oxidized state. Source: (natural)  Methanothermococcus thermolithotrophicus DSM 2095 (archaea) Methanothermococcus thermolithotrophicus DSM 2095 (archaea)Cell line: / / Organ: / / Plasmid details: / / Variant: / / Strain: DSM 2095 / Tissue: / / References: hydroxylamine reductase |
|---|
-Non-polymers , 14 types, 666 molecules 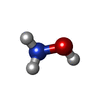






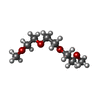



















| #2: Chemical | ChemComp-HOA / #3: Chemical | ChemComp-FMT / #4: Chemical | ChemComp-MG / | #5: Chemical | ChemComp-SF4 / | #6: Chemical | ChemComp-SF3 / | #7: Chemical | ChemComp-PEG / | #8: Chemical | ChemComp-EDO / #9: Chemical | ChemComp-PG5 / | #10: Chemical | ChemComp-GOL / | #11: Chemical | ChemComp-TRS / | #12: Chemical | ChemComp-VQ8 / | #13: Chemical | ChemComp-ACT / #14: Chemical | ChemComp-MRD / ( | #15: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.41 Å3/Da / Density % sol: 49.06 % Description: Brown orthorhombic rod. Appeared after few weeks. |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, sitting drop / pH: 7.6 Details: Crystallisation was performed anaerobically by initial screening at 20 degree Celsius using the sitting drop method on 96-Well MRC 2-Drop polystyrene Crystallisation Plates (SWISSCI) in a ...Details: Crystallisation was performed anaerobically by initial screening at 20 degree Celsius using the sitting drop method on 96-Well MRC 2-Drop polystyrene Crystallisation Plates (SWISSCI) in a Coy tent containing a N2/H2 (97:3%) atmosphere. The reservoir chamber was filled with 90 ul of crystallisation condition, and the crystallisation drop was formed by spotting 0.55 ul protein with 0.55 ul of 20% (w/v) PEG 3,350 and 200 mM Magnesium formate. The protein was crystallised at 9.9 mg/ml in 25 mM Tris/HCl pH 7.6, 2 mM dithiothreitol, 10% (v/v) glycerol. Densities in the electron density map suggest a contamination of another crystallisation condition spatially close and containing 30% (v/v) 2-Methyl-2,4-pentanediol, 20 mM Calcium chloride and 100 mM Sodium acetate, pH 4.6. The crystal was soaked for 5.7 min in a solution of 100 mM hydroxylamine/HCl in the crystallisation condition and then soaked in the crystallisation solution supplemented with 20% v/v ethylene glycol for a few seconds before freezing in liquid nitrogen. PH range: / / Temp details: / |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  PETRA III, DESY PETRA III, DESY  / Beamline: P11 / Wavelength: 1.0004 Å / Beamline: P11 / Wavelength: 1.0004 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Jun 7, 2020 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0004 Å / Relative weight: 1 |
| Reflection | Resolution: 1.356→70.645 Å / Num. obs: 89372 / % possible obs: 94.9 % / Redundancy: 7.1 % / CC1/2: 0.999 / Rmerge(I) obs: 0.11 / Rpim(I) all: 0.043 / Rrim(I) all: 0.118 / Net I/σ(I): 11.5 |
| Reflection shell | Resolution: 1.356→1.476 Å / Redundancy: 5.9 % / Rmerge(I) obs: 1.061 / Mean I/σ(I) obs: 1.6 / Num. unique obs: 4469 / CC1/2: 0.611 / Rpim(I) all: 0.472 / Rrim(I) all: 1.165 / % possible all: 67.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.36→51.09 Å / SU ML: 0.1 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 18.74 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.36→51.09 Å / SU ML: 0.1 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 18.74 / Stereochemistry target values: MLDetails: Refinement steps were performed by considering all atoms anisotropic. The model was refined with hydrogens in riding position. Hydrogens were omitted in the final deposited model.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 18.44 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.36→51.09 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj












