[English] 日本語
 Yorodumi
Yorodumi- PDB-8c9g: Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8c9g | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with mupirocin | ||||||||||||
 Components Components | Isoleucine--tRNA ligase | ||||||||||||
 Keywords Keywords | RNA BINDING PROTEIN / antibiotic / mupirocin-sensitive / IleRS1 | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationisoleucine-tRNA ligase / isoleucyl-tRNA aminoacylation / isoleucine-tRNA ligase activity / aminoacyl-tRNA deacylase activity / tRNA binding / zinc ion binding / ATP binding / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Priestia megaterium (bacteria) Priestia megaterium (bacteria) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | ||||||||||||
 Authors Authors | Brkic, A. / Leibundgut, M. / Jablonska, J. / Zanki, V. / Car, Z. / Petrovic Perokovic, V. / Ban, N. / Gruic-Sovulj, I. | ||||||||||||
| Funding support |  Croatia, Croatia,  Switzerland, European Union, 3items Switzerland, European Union, 3items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif. Authors: Brkic, A. / Leibundgut, M. / Jablonska, J. / Zanki, V. / Car, Z. / Petrovic Perokovic, V. / Marsavelski, A. / Ban, N. / Gruic-Sovulj, I. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8c9g.cif.gz 8c9g.cif.gz | 466.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8c9g.ent.gz pdb8c9g.ent.gz | 302.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8c9g.json.gz 8c9g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c9/8c9g https://data.pdbj.org/pub/pdb/validation_reports/c9/8c9g ftp://data.pdbj.org/pub/pdb/validation_reports/c9/8c9g ftp://data.pdbj.org/pub/pdb/validation_reports/c9/8c9g | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8c8uC  8c8vC  8c8wC  8c9dC  8c9eC  8c9fC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 105090.719 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: Gram-positive bacterium / Source: (gene. exp.)  Priestia megaterium (bacteria) / Strain: DSM-32 / Cell: bacterial / Gene: ileS, BG04_1180 / Variant: de Bary 1884 / Plasmid: pET28b(+) / Details (production host): EMD Biosciences, Cat: 69865-3 / Cell (production host): bacterial / Production host: Priestia megaterium (bacteria) / Strain: DSM-32 / Cell: bacterial / Gene: ileS, BG04_1180 / Variant: de Bary 1884 / Plasmid: pET28b(+) / Details (production host): EMD Biosciences, Cat: 69865-3 / Cell (production host): bacterial / Production host:  |
|---|
-Non-polymers , 6 types, 39 molecules 
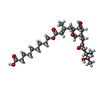









| #2: Chemical | | #3: Chemical | #4: Chemical | ChemComp-CL / #5: Chemical | #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.47 Å3/Da / Density % sol: 50.27 % Description: Plate clusters. Pieces can be broken of the cluster for freezing and diffraction experiments. |
|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: Solution of 20 mg/ml wt-HMGH-PmIleRS1 with added 3 mM mupirocin was mixed in 1:1 ratio with the well solution containing: 400 - 900 mM LiCl and 10 - 20% PEG3350, 25 mM Hepes-KOH pH=7.5 at 20 ...Details: Solution of 20 mg/ml wt-HMGH-PmIleRS1 with added 3 mM mupirocin was mixed in 1:1 ratio with the well solution containing: 400 - 900 mM LiCl and 10 - 20% PEG3350, 25 mM Hepes-KOH pH=7.5 at 20 oC, 50 mM NaCl. The drop volume of 2 microliters, was equilibrated towards 300 microliters of the well solution at 19 degrees. PH range: 6.8-7.5 / Temp details: Crystals do not grow at 4 degrees. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 0.9999873570222 Å / Beamline: X06SA / Wavelength: 0.9999873570222 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Mar 15, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9999873570222 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→60.39 Å / Num. obs: 49643 / % possible obs: 90.26 % / Redundancy: 3.6 % / Biso Wilson estimate: 57.17 Å2 / CC1/2: 0.993 / CC star: 0.998 / Rmerge(I) obs: 0.15 / Rpim(I) all: 0.09249 / Rrim(I) all: 0.1767 / Net I/σ(I): 6.95 |
| Reflection shell | Resolution: 2.8→2.9 Å / Redundancy: 3.6 % / Rmerge(I) obs: 1.288 / Mean I/σ(I) obs: 0.94 / Num. unique obs: 3810 / CC1/2: 0.373 / Rpim(I) all: 0.7865 / Rrim(I) all: 1.513 / % possible all: 75.64 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.8→60.39 Å / SU ML: 0.3967 / Cross valid method: FREE R-VALUE / σ(F): 0.01 / Phase error: 27.9342 MOLECULAR REPLACEMENT / Resolution: 2.8→60.39 Å / SU ML: 0.3967 / Cross valid method: FREE R-VALUE / σ(F): 0.01 / Phase error: 27.9342 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 64.61 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→60.39 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj





