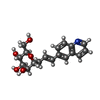
Entry Database : PDB / ID : 8bvdTitle FimH lectin domain in complex with mannose C-linked to quinoline Type 1 fimbrin D-mannose specific adhesin Keywords / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / Biological species Escherichia coli UTI89 (bacteria)Method / / / Resolution : 2.995 Å Authors Bouckaert, J. / Bridot, C. Funding support Organization Grant number Country Centre National de la Recherche Scientifique (CNRS) Natural Sciences and Engineering Research Council (NSERC, Canada) RGPIN-2018-05570
Journal : Pharmaceutics / Year : 2023Title : Insightful Improvement in the Design of Potent Uropathogenic E. coli FimH Antagonists.
Authors :
Mousavifar, L. / Sarshar, M. / Bridot, C. / Scribano, D. / Ambrosi, C. / Palamara, A.T. / Vergoten, G. / Roubinet, B. / Landemarre, L. / Bouckaert, J. / Roy, R. #1: Journal : Molecules / Year : 2017Title : Sites for Dynamic Protein-Carbohydrate Interactions of O- and C-Linked Mannosides on the E. coli FimH Adhesin.
Authors :
Touaibia, M. / Krammer, E.M. / Shiao, T.C. / Yamakawa, N. / Wang, Q. / Glinschert, A. / Papadopoulos, A. / Mousavifar, L. / Maes, E. / Oscarson, S. / Vergoten, G. / Lensink, M.F. / Roy, R. / Bouckaert, J. #2: Journal : Molecules / Year : 2018Title : A Novel Integrated Way for Deciphering the Glycan Code for the FimH Lectin.
Authors :
Dumych, T. / Bridot, C. / Gouin, S.G. / Lensink, M.F. / Paryzhak, S. / Szunerits, S. / Blossey, R. / Bilyy, R. / Bouckaert, J. / Krammer, E.M. History Deposition Dec 3, 2022 Deposition site / Processing site Revision 1.0 Feb 15, 2023 Provider / Type Revision 1.1 Mar 8, 2023 Group / Category Item _citation.journal_abbrev / _citation.journal_id_CSD ... _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_volume / _citation.pdbx_database_id_PubMed / _citation.title Revision 1.2 Nov 6, 2024 Group Data collection / Database references ... Data collection / Database references / Refinement description / Structure summary Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / citation / pdbx_entry_details / pdbx_modification_feature / struct_ncs_dom_lim Item _citation.country / _citation.journal_id_ISSN ... _citation.country / _citation.journal_id_ISSN / _pdbx_entry_details.has_protein_modification / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.995 Å
MOLECULAR REPLACEMENT / Resolution: 2.995 Å  Authors
Authors France,
France,  Canada, 2items
Canada, 2items  Citation
Citation Journal: Pharmaceutics / Year: 2023
Journal: Pharmaceutics / Year: 2023 Journal: Molecules / Year: 2017
Journal: Molecules / Year: 2017 Journal: Molecules / Year: 2018
Journal: Molecules / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 8bvd.cif.gz
8bvd.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb8bvd.ent.gz
pdb8bvd.ent.gz PDB format
PDB format 8bvd.json.gz
8bvd.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/bv/8bvd
https://data.pdbj.org/pub/pdb/validation_reports/bv/8bvd ftp://data.pdbj.org/pub/pdb/validation_reports/bv/8bvd
ftp://data.pdbj.org/pub/pdb/validation_reports/bv/8bvd F&H Search
F&H Search Links
Links Assembly
Assembly




 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SOLEIL
SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.978565 Å
/ Beamline: PROXIMA 1 / Wavelength: 0.978565 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.995→49.732 Å / Cor.coef. Fo:Fc: 0.881 / Cor.coef. Fo:Fc free: 0.83 / WRfactor Rfree: 0.243 / WRfactor Rwork: 0.206 / SU B: 17.437 / SU ML: 0.302 / Average fsc free: 0.9258 / Average fsc work: 0.9484 / Cross valid method: THROUGHOUT / ESU R: 0.669 / ESU R Free: 0.41
MOLECULAR REPLACEMENT / Resolution: 2.995→49.732 Å / Cor.coef. Fo:Fc: 0.881 / Cor.coef. Fo:Fc free: 0.83 / WRfactor Rfree: 0.243 / WRfactor Rwork: 0.206 / SU B: 17.437 / SU ML: 0.302 / Average fsc free: 0.9258 / Average fsc work: 0.9484 / Cross valid method: THROUGHOUT / ESU R: 0.669 / ESU R Free: 0.41  Movie
Movie Controller
Controller



 PDBj
PDBj