+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8ae7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The strucuture of Compound 15 bound to CK2alpha | |||||||||
 Components Components | Casein kinase II subunit alpha | |||||||||
 Keywords Keywords | TRANSFERASE / Fragment based drug discovery | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of chromosome separation / positive regulation of aggrephagy / WNT mediated activation of DVL / Condensation of Prometaphase Chromosomes / protein kinase CK2 complex / symbiont-mediated disruption of host cell PML body / Receptor Mediated Mitophagy / Synthesis of PC / Sin3-type complex / Maturation of hRSV A proteins ...regulation of chromosome separation / positive regulation of aggrephagy / WNT mediated activation of DVL / Condensation of Prometaphase Chromosomes / protein kinase CK2 complex / symbiont-mediated disruption of host cell PML body / Receptor Mediated Mitophagy / Synthesis of PC / Sin3-type complex / Maturation of hRSV A proteins / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / negative regulation of signal transduction by p53 class mediator / negative regulation of apoptotic signaling pathway / positive regulation of Wnt signaling pathway / negative regulation of double-strand break repair via homologous recombination / : / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / Signal transduction by L1 / Hsp90 protein binding / PML body / Regulation of PTEN stability and activity / Wnt signaling pathway / positive regulation of protein catabolic process / kinase activity / KEAP1-NFE2L2 pathway / rhythmic process / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / double-strand break repair / positive regulation of cell growth / Regulation of TP53 Activity through Phosphorylation / non-specific serine/threonine protein kinase / regulation of cell cycle / negative regulation of translation / protein stabilization / protein serine kinase activity / protein serine/threonine kinase activity / positive regulation of cell population proliferation / apoptotic process / DNA damage response / signal transduction / nucleoplasm / ATP binding / identical protein binding / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.28 Å molecular replacement / Resolution: 1.28 Å | |||||||||
 Authors Authors | Brear, P. / De Fusco, C. / Atkinson, E. / Frances, N. / Iegre, J. / Venkitaraman, A. / Hyvonen, M. / Spring, D. | |||||||||
| Funding support |  United Kingdom, 2items United Kingdom, 2items
| |||||||||
 Citation Citation |  Journal: Rsc Med Chem / Year: 2022 Journal: Rsc Med Chem / Year: 2022Title: A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action. Authors: Brear, P. / De Fusco, C. / Atkinson, E.L. / Iegre, J. / Francis-Newton, N.J. / Venkitaraman, A.R. / Hyvonen, M. / Spring, D.R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8ae7.cif.gz 8ae7.cif.gz | 95.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8ae7.ent.gz pdb8ae7.ent.gz | 69.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8ae7.json.gz 8ae7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ae/8ae7 https://data.pdbj.org/pub/pdb/validation_reports/ae/8ae7 ftp://data.pdbj.org/pub/pdb/validation_reports/ae/8ae7 ftp://data.pdbj.org/pub/pdb/validation_reports/ae/8ae7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7zy0C  7zy2C  7zy5C  7zy8C  7zydC  7zykC  7zyoC  7zyrC  8aecC  8aekC  8aemC  3pe1S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 39031.391 Da / Num. of mol.: 1 / Mutation: K74A, K75A, K76A, R21S Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CSNK2A1, CK2A1 / Production host: Homo sapiens (human) / Gene: CSNK2A1, CK2A1 / Production host:  References: UniProt: P68400, non-specific serine/threonine protein kinase |
|---|
-Non-polymers , 6 types, 344 molecules 

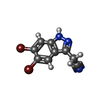








| #2: Chemical | ChemComp-GOL / |
|---|---|
| #3: Chemical | ChemComp-ACT / |
| #4: Chemical | ChemComp-LVU / |
| #5: Chemical | ChemComp-ADP / |
| #6: Chemical | ChemComp-MG / |
| #7: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.03 Å3/Da / Density % sol: 39.49 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 112.5mM Mes pH 6.5, 35% glycerol ethoxylate, 180 mM ammonium acetate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9184 Å / Beamline: I04 / Wavelength: 0.9184 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Sep 16, 2016 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9184 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.28→59.02 Å / Num. obs: 80979 / % possible obs: 100 % / Redundancy: 6.4 % / Biso Wilson estimate: 12.98 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.074 / Rpim(I) all: 0.032 / Rrim(I) all: 0.081 / Net I/σ(I): 10.3 / Num. measured all: 517476 / Scaling rejects: 628 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3PE1 Resolution: 1.28→54.47 Å / Cor.coef. Fo:Fc: 0.9601 / Cor.coef. Fo:Fc free: 0.9493 / SU R Cruickshank DPI: 0.057 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.058 / SU Rfree Blow DPI: 0.06 / SU Rfree Cruickshank DPI: 0.059
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 107.38 Å2 / Biso mean: 20.2 Å2 / Biso min: 5.18 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.196 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.28→54.47 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.28→1.31 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller



 PDBj
PDBj









