+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7up3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | NDM1-inhibitor co-structure | ||||||
 Components Components | Metallo beta-lactamase | ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / Metallo-beta-lactamase inhibitor / HYDROLASE / HYDROLASE-HYDROLASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 1.4 Å FOURIER SYNTHESIS / Resolution: 1.4 Å | ||||||
 Authors Authors | Scapin, G. / Fischmann, T.O. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2022 Journal: J.Med.Chem. / Year: 2022Title: Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates. Authors: Mandal, M. / Xiao, L. / Pan, W. / Scapin, G. / Li, G. / Tang, H. / Yang, S.W. / Pan, J. / Root, Y. / de Jesus, R.K. / Yang, C. / Prosise, W. / Dayananth, P. / Mirza, A. / Therien, A.G. / ...Authors: Mandal, M. / Xiao, L. / Pan, W. / Scapin, G. / Li, G. / Tang, H. / Yang, S.W. / Pan, J. / Root, Y. / de Jesus, R.K. / Yang, C. / Prosise, W. / Dayananth, P. / Mirza, A. / Therien, A.G. / Young, K. / Flattery, A. / Garlisi, C. / Zhang, R. / Chu, D. / Sheth, P. / Chu, I. / Wu, J. / Markgraf, C. / Kim, H.Y. / Painter, R. / Mayhood, T.W. / DiNunzio, E. / Wyss, D.F. / Buevich, A.V. / Fischmann, T. / Pasternak, A. / Dong, S. / Hicks, J.D. / Villafania, A. / Liang, L. / Murgolo, N. / Black, T. / Hagmann, W.K. / Tata, J. / Parmee, E.R. / Weber, A.E. / Su, J. / Tang, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7up3.cif.gz 7up3.cif.gz | 192.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7up3.ent.gz pdb7up3.ent.gz | 148.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7up3.json.gz 7up3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7up3_validation.pdf.gz 7up3_validation.pdf.gz | 1.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7up3_full_validation.pdf.gz 7up3_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  7up3_validation.xml.gz 7up3_validation.xml.gz | 22.2 KB | Display | |
| Data in CIF |  7up3_validation.cif.gz 7up3_validation.cif.gz | 32 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/up/7up3 https://data.pdbj.org/pub/pdb/validation_reports/up/7up3 ftp://data.pdbj.org/pub/pdb/validation_reports/up/7up3 ftp://data.pdbj.org/pub/pdb/validation_reports/up/7up3 | HTTPS FTP |
-Related structure data
| Related structure data |  7uoxC  7uoyC  7up1C  7up2C  3zr9S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 26294.578 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria)Gene: blaNDM-1, bla NDM-1, blaNDM1, NDM-1, D647_p47098, DDJ63_29735, EC13450_007, NCTC13443_00040, p2146_00143, pCRE380_21, pN11x00042NDM_090, pNDM-SX04_5, pNDM10469_138, SAMEA3499901_05193, TR3_031, TR4_031 Production host:  |
|---|
-Non-polymers , 5 types, 322 molecules 

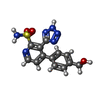






| #2: Chemical | ChemComp-CD / #3: Chemical | #4: Chemical | #5: Chemical | ChemComp-EPE / | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.89 Å3/Da / Density % sol: 34.92 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 0.2 M ammonium acetate 1.0 mM TCEP 3.0 mM cadmium sulfate 0.1 M HEPES pH 7.0 25.0 w/v polyethylene glycol 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 Å / Beamline: 17-ID / Wavelength: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Feb 22, 2013 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.113→134.175 Å / Num. all: 163291 / Num. obs: 163291 / % possible obs: 100 % / Redundancy: 12.8 % / Biso Wilson estimate: 17.2 Å2 / Rpim(I) all: 0.029 / Rrim(I) all: 0.105 / Rsym value: 0.094 / Net I/av σ(I): 5.9 / Net I/σ(I): 18 / Num. measured all: 2088252 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / % possible all: 100
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: 3ZR9 Resolution: 1.4→31.72 Å / Cor.coef. Fo:Fc: 0.9434 / Cor.coef. Fo:Fc free: 0.9418 / SU R Cruickshank DPI: 0.063 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.065 / SU Rfree Blow DPI: 0.061 / SU Rfree Cruickshank DPI: 0.06
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 106.58 Å2 / Biso mean: 23.06 Å2 / Biso min: 10.04 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.171 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.4→31.72 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.4→1.44 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj





