[English] 日本語
 Yorodumi
Yorodumi- PDB-7ufv: Crystal structure of the WDR domain of human DCAF1 in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7ufv | ||||||
|---|---|---|---|---|---|---|---|
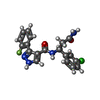
| Title | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | ||||||
 Components Components | DDB1- and CUL4-associated factor 1 | ||||||
 Keywords Keywords | TRANSFERASE / WD-repeat / WDR / DCAF1 / SGC / Structural Genomics / Structural Genomics Consortium | ||||||
| Function / homology |  Function and homology information Function and homology informationcell competition in a multicellular organism / histone H2AT120 kinase activity / V(D)J recombination / Cul4-RING E3 ubiquitin ligase complex / ubiquitin-like ligase-substrate adaptor activity / post-translational protein modification / B cell differentiation / nuclear estrogen receptor binding / fibrillar center / positive regulation of protein catabolic process ...cell competition in a multicellular organism / histone H2AT120 kinase activity / V(D)J recombination / Cul4-RING E3 ubiquitin ligase complex / ubiquitin-like ligase-substrate adaptor activity / post-translational protein modification / B cell differentiation / nuclear estrogen receptor binding / fibrillar center / positive regulation of protein catabolic process / Antigen processing: Ubiquitination & Proteasome degradation / proteasome-mediated ubiquitin-dependent protein catabolic process / non-specific serine/threonine protein kinase / protein ubiquitination / protein serine kinase activity / centrosome / negative regulation of transcription by RNA polymerase II / nucleoplasm / ATP binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 1.9 Å FOURIER SYNTHESIS / Resolution: 1.9 Å | ||||||
 Authors Authors | Kimani, S. / Li, A. / Li, Y. / Dong, A. / Hutchinson, A. / Seitova, A. / Wilson, B. / Al-Awar, R. / Vedadi, M. / Brown, P. ...Kimani, S. / Li, A. / Li, Y. / Dong, A. / Hutchinson, A. / Seitova, A. / Wilson, B. / Al-Awar, R. / Vedadi, M. / Brown, P. / Arrowsmith, C.H. / Edwards, A.M. / Halabelian, L. / Structural Genomics Consortium (SGC) | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2023 Journal: J.Med.Chem. / Year: 2023Title: Discovery of Nanomolar DCAF1 Small Molecule Ligands. Authors: Li, A.S.M. / Kimani, S. / Wilson, B. / Noureldin, M. / Gonzalez-Alvarez, H. / Mamai, A. / Hoffer, L. / Guilinger, J.P. / Zhang, Y. / von Rechenberg, M. / Disch, J.S. / Mulhern, C.J. / ...Authors: Li, A.S.M. / Kimani, S. / Wilson, B. / Noureldin, M. / Gonzalez-Alvarez, H. / Mamai, A. / Hoffer, L. / Guilinger, J.P. / Zhang, Y. / von Rechenberg, M. / Disch, J.S. / Mulhern, C.J. / Slakman, B.L. / Cuozzo, J.W. / Dong, A. / Poda, G. / Mohammed, M. / Saraon, P. / Mittal, M. / Modh, P. / Rathod, V. / Patel, B. / Ackloo, S. / Santhakumar, V. / Szewczyk, M.M. / Barsyte-Lovejoy, D. / Arrowsmith, C.H. / Marcellus, R. / Guie, M.A. / Keefe, A.D. / Brown, P.J. / Halabelian, L. / Al-Awar, R. / Vedadi, M. #1:  Journal: J.Chem.Inf.Model. / Year: 2023 Journal: J.Chem.Inf.Model. / Year: 2023Title: Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets. Authors: Kimani, S.W. / Owen, J. / Green, S.R. / Li, F. / Li, Y. / Dong, A. / Brown, P.J. / Ackloo, S. / Kuter, D. / Yang, C. / MacAskill, M. / MacKinnon, S.S. / Arrowsmith, C.H. / Schapira, M. / ...Authors: Kimani, S.W. / Owen, J. / Green, S.R. / Li, F. / Li, Y. / Dong, A. / Brown, P.J. / Ackloo, S. / Kuter, D. / Yang, C. / MacAskill, M. / MacKinnon, S.S. / Arrowsmith, C.H. / Schapira, M. / Shahani, V. / Halabelian, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7ufv.cif.gz 7ufv.cif.gz | 131.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7ufv.ent.gz pdb7ufv.ent.gz | 99.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7ufv.json.gz 7ufv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uf/7ufv https://data.pdbj.org/pub/pdb/validation_reports/uf/7ufv ftp://data.pdbj.org/pub/pdb/validation_reports/uf/7ufv ftp://data.pdbj.org/pub/pdb/validation_reports/uf/7ufv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7sseS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 35592.945 Da / Num. of mol.: 2 / Mutation: F1077A, R1079A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: DCAF1, KIAA0800, RIP, VPRBP / Plasmid: pFBOH-MHL / Cell line (production host): Sf9 / Production host: Homo sapiens (human) / Gene: DCAF1, KIAA0800, RIP, VPRBP / Plasmid: pFBOH-MHL / Cell line (production host): Sf9 / Production host:  References: UniProt: Q9Y4B6, non-specific serine/threonine protein kinase #2: Chemical | #3: Chemical | ChemComp-UNX / | #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.21 Å3/Da / Density % sol: 44.44 % / Mosaicity: 0.18 ° |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 7.4 / Details: 20% PEG3350, 0.2M Potassium Fluoride |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-C / Wavelength: 0.9791 Å / Beamline: 24-ID-C / Wavelength: 0.9791 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER2 X 16M / Detector: PIXEL / Date: Oct 24, 2021 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9791 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.9→43.98 Å / Num. obs: 48379 / % possible obs: 99 % / Redundancy: 3.8 % / CC1/2: 0.998 / Rmerge(I) obs: 0.062 / Rpim(I) all: 0.036 / Rrim(I) all: 0.072 / Net I/σ(I): 9.6 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: 7SSE Resolution: 1.9→43.98 Å / Cor.coef. Fo:Fc: 0.965 / Cor.coef. Fo:Fc free: 0.943 / SU B: 6.386 / SU ML: 0.167 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.164 / ESU R Free: 0.157 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 42.3 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→43.98 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj