[English] 日本語
 Yorodumi
Yorodumi- PDB-7ubi: Solution NMR structure of 8-residue Rosetta-designed cyclic pepti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7ubi | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution NMR structure of 8-residue Rosetta-designed cyclic peptide D8.21 in CDCl3 with cis/trans switching (TT conformation, 47%) | ||||||
 Components Components | Cyclic peptide D8.21 DVA-MLE-DPR-LEU-DVA-MLE-DPR-LEU | ||||||
 Keywords Keywords | DE NOVO PROTEIN / cyclic peptide / non natural amino acids / cis/trans / switch peptides / de novo design / membrane permeability | ||||||
| Function / homology | polypeptide(D) Function and homology information Function and homology information | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method | SOLUTION NMR / na | ||||||
 Authors Authors | Ramelot, T.A. / Tejero, R. / Montelione, G.T. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Accurate de novo design of membrane-traversing macrocycles. Authors: Bhardwaj, G. / O'Connor, J. / Rettie, S. / Huang, Y.H. / Ramelot, T.A. / Mulligan, V.K. / Alpkilic, G.G. / Palmer, J. / Bera, A.K. / Bick, M.J. / Di Piazza, M. / Li, X. / Hosseinzadeh, P. / ...Authors: Bhardwaj, G. / O'Connor, J. / Rettie, S. / Huang, Y.H. / Ramelot, T.A. / Mulligan, V.K. / Alpkilic, G.G. / Palmer, J. / Bera, A.K. / Bick, M.J. / Di Piazza, M. / Li, X. / Hosseinzadeh, P. / Craven, T.W. / Tejero, R. / Lauko, A. / Choi, R. / Glynn, C. / Dong, L. / Griffin, R. / van Voorhis, W.C. / Rodriguez, J. / Stewart, L. / Montelione, G.T. / Craik, D. / Baker, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7ubi.cif.gz 7ubi.cif.gz | 57.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7ubi.ent.gz pdb7ubi.ent.gz | 41.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7ubi.json.gz 7ubi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ub/7ubi https://data.pdbj.org/pub/pdb/validation_reports/ub/7ubi ftp://data.pdbj.org/pub/pdb/validation_reports/ub/7ubi ftp://data.pdbj.org/pub/pdb/validation_reports/ub/7ubi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7ubcC  7ubdC  7ubeC  7ubfC  7ubgC  7ubhC  7ucpC  7uzlC  8ctoC  8cunC  8cwaC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 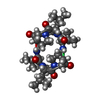
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Polypeptide(D) | Mass: 891.190 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|---|
| Has ligand of interest | N |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution / Contents: 6 mg/mL peptide, 0.03 % TMS, CDCl3 / Label: D8.21-CDCl3 / Solvent system: CDCl3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||
| Sample conditions | Ionic strength: NA Not defined / Label: conditions_1 / pH: 0.0 Not defined / Pressure: 1 atm / Temperature: 293 K / Temperature err: 1 |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE III / Manufacturer: Bruker / Model: AVANCE III / Field strength: 600 MHz / Details: CP TCI - Pharaoh - RPI |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: na / Software ordinal: 1 Details: This is conformation A (TT) of the same 8-residue cyclic peptide present in the same NMR sample. TT symmetric conformation has trans DPR3 and trans DPR7 (47%). TC conformation has trans DPR3 and cis DPR7 (53%). | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: medoid | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller


 PDBj
PDBj
 HSQC
HSQC