+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 7s51 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
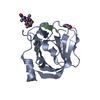
| タイトル | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATA peptide | |||||||||
 要素 要素 |
| |||||||||
 キーワード キーワード | HYDROLASE / sortase-fold / sortase / eight-stranded beta barrel / transpeptidase / housekeeping sortase / surface protein | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報 | |||||||||
| 生物種 |  Streptococcus pyogenes (化膿レンサ球菌) Streptococcus pyogenes (化膿レンサ球菌)Synthetic construct (人工物) | |||||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 1.4 Å 分子置換 / 解像度: 1.4 Å | |||||||||
 データ登録者 データ登録者 | Johnson, D.A. / Svendsen, J.E. / Antos, J.M. / Amacher, J.F. | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: J.Biol.Chem. / 年: 2022 ジャーナル: J.Biol.Chem. / 年: 2022タイトル: Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition. 著者: Johnson, D.A. / Piper, I.M. / Vogel, B.A. / Jackson, S.N. / Svendsen, J.E. / Kodama, H.M. / Lee, D.E. / Lindblom, K.M. / McCarty, J. / Antos, J.M. / Amacher, J.F. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  7s51.cif.gz 7s51.cif.gz | 87.6 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb7s51.ent.gz pdb7s51.ent.gz | 64.1 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  7s51.json.gz 7s51.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  7s51_validation.pdf.gz 7s51_validation.pdf.gz | 452.7 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  7s51_full_validation.pdf.gz 7s51_full_validation.pdf.gz | 458.8 KB | 表示 | |
| XML形式データ |  7s51_validation.xml.gz 7s51_validation.xml.gz | 17.9 KB | 表示 | |
| CIF形式データ |  7s51_validation.cif.gz 7s51_validation.cif.gz | 25.6 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/s5/7s51 https://data.pdbj.org/pub/pdb/validation_reports/s5/7s51 ftp://data.pdbj.org/pub/pdb/validation_reports/s5/7s51 ftp://data.pdbj.org/pub/pdb/validation_reports/s5/7s51 | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 単位格子 |
|
- 要素
要素
| #1: タンパク質 | 分子量: 18589.051 Da / 分子数: 2 / 変異: C208A / 由来タイプ: 組換発現 由来: (組換発現)  Streptococcus pyogenes (化膿レンサ球菌) Streptococcus pyogenes (化膿レンサ球菌)遺伝子: srtA, srtA_1, srtA_2, E0F66_05345, E0F67_00760, FGO82_09960, FNL90_04725, FNL91_04720, GQ677_05600, GQR49_04420, GQY31_04460, GQY92_04850, GTK43_04765, GTK52_04270, GTK53_04530, GTK54_ ...遺伝子: srtA, srtA_1, srtA_2, E0F66_05345, E0F67_00760, FGO82_09960, FNL90_04725, FNL91_04720, GQ677_05600, GQR49_04420, GQY31_04460, GQY92_04850, GTK43_04765, GTK52_04270, GTK53_04530, GTK54_03910, GUA39_04435, IB935_04675, IB936_04605, IB937_04535, IB938_05195, KUN2590_09100, KUN4944_08330, MGAS2221_0893, SAMEA1407055_00305, SAMEA1711644_00960, SAMEA3918953_00457, SPNIH34_10200, SPNIH35_09070 プラスミド: pET28a(+) / 発現宿主:  #2: タンパク質・ペプチド | 分子量: 864.000 Da / 分子数: 2 / 由来タイプ: 合成 / 由来: (合成) Synthetic construct (人工物) #3: 水 | ChemComp-HOH / | 研究の焦点であるリガンドがあるか | N | |
|---|
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 1.92 Å3/Da / 溶媒含有率: 35.91 % |
|---|---|
| 結晶化 | 温度: 298 K / 手法: 蒸気拡散法, ハンギングドロップ法 / pH: 6 詳細: 0.1 M Tris pH 6, 34% (w/v) PEG 8000, 0.1 M sodium acetate |
-データ収集
| 回折 | 平均測定温度: 80 K / Serial crystal experiment: N | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  ALS ALS  / ビームライン: 5.0.1 / 波長: 0.97741 Å / ビームライン: 5.0.1 / 波長: 0.97741 Å | ||||||||||||||||||||||||
| 検出器 | タイプ: DECTRIS PILATUS3 6M / 検出器: PIXEL / 日付: 2020年12月12日 | ||||||||||||||||||||||||
| 放射 | モノクロメーター: Si(220) / プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray | ||||||||||||||||||||||||
| 放射波長 | 波長: 0.97741 Å / 相対比: 1 | ||||||||||||||||||||||||
| 反射 | 解像度: 1.4→48.94 Å / Num. obs: 57131 / % possible obs: 99.9 % / 冗長度: 12.6 % / Biso Wilson estimate: 10.34 Å2 / CC1/2: 0.998 / Rmerge(I) obs: 0.097 / Rpim(I) all: 0.028 / Rrim(I) all: 0.101 / Net I/σ(I): 13.6 / Num. measured all: 719759 / Scaling rejects: 4283 | ||||||||||||||||||||||||
| 反射 シェル | Diffraction-ID: 1
|
- 解析
解析
| ソフトウェア |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法:  分子置換 分子置換開始モデル: 3FN5 解像度: 1.4→48.937 Å / SU ML: 0.15 / 交差検証法: THROUGHOUT / σ(F): 1.34 / 位相誤差: 26.51 / 立体化学のターゲット値: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | 減衰半径: 0.9 Å / VDWプローブ半径: 1.11 Å / 溶媒モデル: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso max: 59.65 Å2 / Biso mean: 14.4023 Å2 / Biso min: 3.41 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: final / 解像度: 1.4→48.937 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS精密化 シェル | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0
|
 ムービー
ムービー コントローラー
コントローラー






 PDBj
PDBj
