登録情報 データベース : PDB / ID : 7r6rタイトル Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3')DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3')Immunity repressor キーワード / / / / / 機能・相同性 / / 生物種 手法 / / / 解像度 : 3.13 Å データ登録者 McGinnis, R.J. / Brambley, C.A. / Stamey, B. / Green, W.C. / Gragg, K.N. / Cafferty, E.R. / Terwilliger, T.C. / Hammel, M. / Hollis, T.J. / Miller, J.M. ...McGinnis, R.J. / Brambley, C.A. / Stamey, B. / Green, W.C. / Gragg, K.N. / Cafferty, E.R. / Terwilliger, T.C. / Hammel, M. / Hollis, T.J. / Miller, J.M. / Gainey, M.D. / Wallen, J.R. 資金援助 組織 認可番号 国 Howard Hughes Medical Institute (HHMI)
ジャーナル : Nat Commun / 年 : 2022タイトル : A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.著者 : McGinnis, R.J. / Brambley, C.A. / Stamey, B. / Green, W.C. / Gragg, K.N. / Cafferty, E.R. / Terwilliger, T.C. / Hammel, M. / Hollis, T.J. / Miller, J.M. / Gainey, M.D. / Wallen, J.R. 履歴 登録 2021年6月23日 登録サイト / 処理サイト 改定 1.0 2022年7月20日 Provider / タイプ 改定 1.1 2022年7月27日 Group / カテゴリ / citation_authorItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID / _citation_author.name 改定 1.2 2023年10月18日 Group / Refinement descriptionカテゴリ / chem_comp_bond / pdbx_initial_refinement_model
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Mycobacterium phage TipsytheTRex (ファージ)
Mycobacterium phage TipsytheTRex (ファージ) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 3.13 Å
分子置換 / 解像度: 3.13 Å  データ登録者
データ登録者 米国, 1件
米国, 1件  引用
引用 ジャーナル: Nat Commun / 年: 2022
ジャーナル: Nat Commun / 年: 2022 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 7r6r.cif.gz
7r6r.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb7r6r.ent.gz
pdb7r6r.ent.gz PDB形式
PDB形式 7r6r.json.gz
7r6r.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 7r6r_validation.pdf.gz
7r6r_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 7r6r_full_validation.pdf.gz
7r6r_full_validation.pdf.gz 7r6r_validation.xml.gz
7r6r_validation.xml.gz 7r6r_validation.cif.gz
7r6r_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/r6/7r6r
https://data.pdbj.org/pub/pdb/validation_reports/r6/7r6r ftp://data.pdbj.org/pub/pdb/validation_reports/r6/7r6r
ftp://data.pdbj.org/pub/pdb/validation_reports/r6/7r6r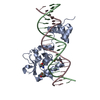
 F&H 検索
F&H 検索 リンク
リンク 集合体
集合体
 要素
要素 Mycobacterium phage TipsytheTRex (ファージ)
Mycobacterium phage TipsytheTRex (ファージ)
 Mycobacterium phage TipsytheTRex (ファージ)
Mycobacterium phage TipsytheTRex (ファージ) Mycobacterium phage TipsytheTRex (ファージ)
Mycobacterium phage TipsytheTRex (ファージ) X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  ALS
ALS  / ビームライン: 8.3.1 / 波長: 1.11608 Å
/ ビームライン: 8.3.1 / 波長: 1.11608 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー



 PDBj
PDBj





































